3845
Residual water signal removal in MR spectroscopic imaging with L2 regularization1Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Department of Electronic Science, Xiamen University, Xiamen, China, 3F. M. Kirby Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Synopsis
Residual water signals in MRSI data may hinder the quantification of metabolite signals and thus affect the quality of final metabolite maps. A L2 regularization based post-processing method is proposed here for efficient removal of residual water signals especially in MRSI data. Using a water-basis matrix, the proposed method aims to find spectra that match the original metabolite signals, but at the same time imposes a constraint of reduced water signals. Results show that the L2 regularization based method can be a highly effective way for removing residual water signals from MRSI data of human brain.
Introduction
Magnetic Resonance Spectroscopic Imaging (MRSI) is a useful tool for noninvasive spatial mapping of tissue metabolites, and has been used to characterize diseases in the human brain, prostate, and breast. In comparison to single voxel MRS experiments, water suppression with pulse sequence methods becomes less efficient and spatially dependent in MRSI experiments due to B0-field inhomogeneity. Insufficient water suppression may lead to bias in metabolite quantification1. Therefore, the residual water signals needs to be removed in post-processing. Several efficient post-processing methods based on singular value decomposition (SVD) or time dimension filters have been reported for the removal of residual water signals for in vivo MR spectroscopy (MRS)2-4. However, these methods generally assume that MRS signals in the time domain are composed of a limited number of damped exponentials, and they may be invalid when the residual water signal (for example, with oscillating tails) do not fit the expected model well. In this abstract, using a set of artificial water-basis signals (orthogonal to metabolite signals in the spectral dimension), an L2 regularization based post-processing method for better removal of residual water signals, especially in MRSI data, is proposed.Methods
The residual water signal can be minimized in the spectral dimension by solving the following optimization problem as
min||x-x0||22+β||WHx||22, (1)
where x0 is the original MR spectrum with residual water signal; W the artificial water-basis matrix; x the optimized metabolite spectrum with water suppressed, and β a regularization parameter. Equation (1) aims to find a spectrum that matches original metabolite signals, but at the same time imposes a constraint of reduced water signal. A simplified closed-form solution for equation (1) can be obtained as
x = (I+β·WWH)-1x0. (2)
This method was compared with conventional Marion2 and HSVD3 methods on simulated signals and in vivo MRSI data. Simulations and data processing were all carried out with in-house programs developed with MATLAB. The parameter β was set to 10-3 for efficient water removal.
For spectral simulations, a set of echo-type signals with different water amplification factors (WF) were generated using
S(t) = ∑k=0mSk(t) = ∑k=0mAke-dk|t-T/2|ei(ωkt+Φk)U(t), (3)
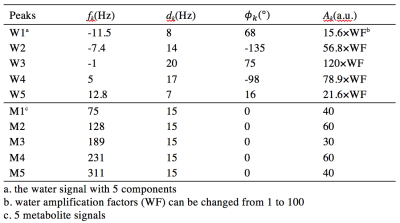
where Ak were amplitudes of signals, dk the damping factors, T the acquisition length, ωk and Φk the signal precession frequencies and phases, and U(t) the Heaviside step function3. These simulated signals (parameters listed in Table 1) were sampled with 512 points at a bandwidth of 2000 Hz (T = 256 ms). For removal of the water signal with L2 regularization, an artificial water-basis matrix W with 1000 water signals were constructed (Figure 1e).
To demonstrate feasibility of the method in vivo, 4-slice spin-echo MRSI data were acquired in a healthy volunteer on a Philips Achieva 3T scanner using a 32-channel head coil (field of view = 230 × 172.5 mm2; slice matrix = 32 × 24; SENSE factor = 2 × 1.5; slice thickness = 15 mm; slice gap = 2.5 mm; echo time = 140 ms; repetition time = 3.2 s; sample bandwidth = 2000 Hz; sample points = 512; 1 transient; acquisition time = 13.5 min). A single chemical shift selective suppression (CHESS) pulse (bandwidth 100 Hz) was applied for water suppression, and the lipid signals were suppressed with octagonal outer volume suppression pulses. Residual lipid signals were further removed in post-processing with L2 regularization5 after removal of residual water signals.
Results and discussion
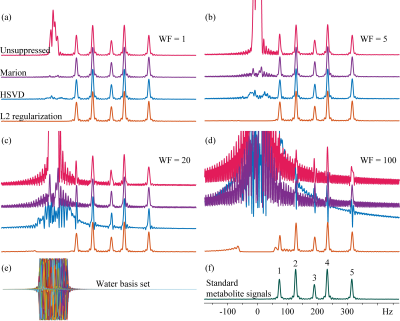
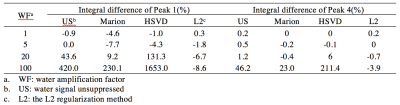
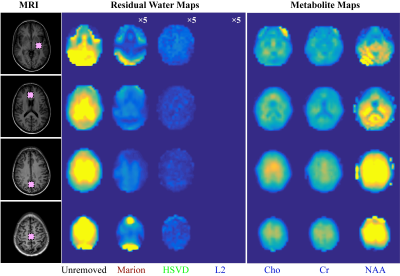
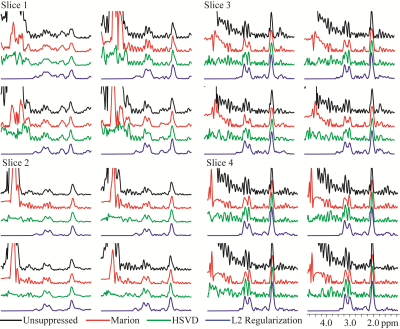
Results of different water removal methods on simulated signals are shown (Figure 1). Differences of peak integrals (1 and 4) from their true values (Figure 1f) under different methods are listed in Table 2. It can be seen that the L2 regularization based water removal method can recover favorable metabolite peaks and spectral baselines. and keep integral differences less than 10% (peak 1, close to the water signal) and 5% (peak 4) under increased water intensities (WF = 20 and 100) where the other two methods (Marion and HSVD) breakdown (Figure 1c, 1d). For the in vivo results, residual water signals along with oscillating tails are well removed by L2 regularization (see residual water maps in Figure 2 and individual spectra in Figure 3), leading to the creation of metabolic images (Figure 2) largely free from artifact due to residual water.
In summary, L2 regularization using a water basis matrix is shown to be a highly effective way of removing residual water signals from MRSI data of the human brain. While L2 regularization has been previously used for lipid suppression in MRSI5, to the best of our knowledge it has not been previously used for water signal removal.
Acknowledgements
Grant sponsor: National Institutes of Health; Grant number: NIH P41EB015909
Liangjie Lin thanks the financial support from China Scholarships Council (No. 201606310173).
References
1 H. Zhu, R. Ouwerkerk, and P. Barker, Magn. Reson. Med. 63, 1486-1492 (2010).
2 D. Marion, M. Ikura, and A. Bax, J. Magn. Reson. 84, 425–430 (1989).
3 H. Barkhuysen, R. de Beer, and D. van Ormondt, J. Magn. Reson. 73, 553–557 (1987).
4 A. Coron, L. Vanhamme, J. Antoine, P. Hecke, and S. Huffel, J. Magn. Reson. 152, 26-40 (2001).
5 B. Bilgic, I. Chatnuntawech, A. Fan, K. Setsompop, S. Cauley, L. Wald, and E. Adalsteinsson, J. Magn. Reson. Imaging 40, 181-191 (2014).
Figures