3840
Over-discretized SENSE reconstruction and B0 correction for accelerated non-lipid suppressed 1H FID MRSI of the human brain at 9.4T1Max Planck Institute for Biological Cybernetics, Tuebingen, Germany, 2IMPRS for Cognitive and Systems Neuroscience, Eberhard-Karls University of Tuebingen, Tuebingen, Germany, 3Institute of Physics, Ernst-Moritz-Arndt University Greifswald, Greifswald, Germany
Synopsis
In this study the acquisition of high resolution (64x64) metabolite maps at 9.4T using a non-lipid suppressed ultra-short TR and TE 1H FID MRSI sequence is accelerated using an improved over-discretized SENSE reconstruction and B0 correction method. The improved reconstruction is compared to conventional SENSE and GRAPPA reconstruction, and reproducible metabolite maps are acquired using this technique.
Introduction
Non-lipid suppressed ultra-short TR and TE 1H FID MRSI 1-4 is a powerful technique for metabolite mapping at ultra-high fields. However, there is still a need for further accelerating this sequence. Parallel imaging techniques can be used for this purpose, however, the subcutaneous lipid signals make the use of parallel imaging methods extremely challenging in this case, as any residual aliasing artifact resulting from an unreliable reconstruction can lead to strong lipid contamination inside the brain.
Recently there was an improved SENSE reconstruction method proposed by Kirchner et al5, 6. This Over-discretized SENSE reconstruction method was shown to reduce voxel bleeding artifacts by accounting for sub-voxel coil sensitivity differences. Additionally, using a high resolution B0 map to correct for the frequency shifts on a sub-voxel level was proven to provide additional SNR gain and line-shape improvement, which is especially beneficial for highly accelerated data.
The benefits of this improved SENSE reconstruction have previously been only demonstrated for the case of lipid-suppressed and lower resolution MRSI5, 6. Therefore, the aim of this work was to explore the benefits of over-discrete SENSE reconstruction along with the B0 correction for accelerating non-lipid suppressed ultra-short TR and TE 1H FID MRSI data at 9.4T, and to perform reliable high resolution metabolite mapping using the highest possible acceleration factor.
Methods
High resolution (3.125x3.125x10mm3) single-slice ultra-short TR and TE non-lipid suppressed 1H FID MRSI1-4 data were acquired from the brains of healthy volunteers on a 9.4T Siemens whole-body human scanner with a TR of 300ms. Other parameters include: FOV = 200x200mm, TE = 1.5ms, BW= 8kHz, in-plane matrix size = 64x64. No lipid or outer volume suppression was used in this sequence. A short (62ms) three-pulse water suppression scheme was incorporated before the excitation pulse. A high resolution scout anatomical image was used to extract the coil sensitivities using the ESPIRiT7 method, and a high resolution B0 map was also acquired using a 2D GRE dual-echo sequence for B0 correction.
The MRSI data was retrospectively undersampled by 7 different acceleration factors and reconstructed with two schemes: 1) conventional SENSE, 2) over-discrete SENSE (OD-SENSE) with an over-discretization factor of 4, along with a Gaussian shape target matrix with the FWHM of the nominal voxel-size in addition to B0 correction on a sub-voxel level. Additionally, the OD-SENSE reconstruction method was compared to a conventional GRAPPA8 reconstruction for the highest reliable acceleration factor.
Finally, the reproducibility of SENSE-accelerated high resolution metabolite mapping was tested on several volunteers.
Results/Discussion
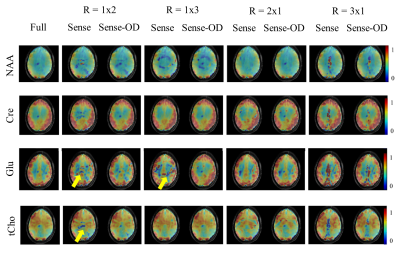
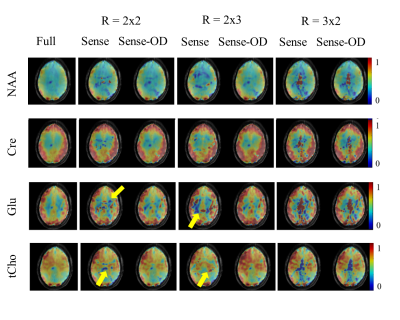
Figure 1 and 2 show metabolite maps of 4 major metabolites for the case of fully sampled data as well as ones accelerated in one (Figure 1) or both phase encoding directions (Figure 2). The accelerated data is shown reconstructed once with the conventional and once with the OD-SENSE reconstruction schemes. It can be seen that the over-discretized reconstruction outperforms conventional SENSE. The advantage of this method can be clearly seen from Figure 1 and 2 since the conventional SENSE results in residual lipid ring aliasing artifacts (e.g. R= 1x2, 1x3 or 2x2).
Figure 3 shows lipid contamination maps along with representative spectra for R=2x2 for the fully sampled as well as the two SENSE reconstruction schemes. These results further highlight the advantage of the improved SENSE reconstruction in reducing the residual lipid contamination artifacts.
The average SNR of the OD-SENSE for R=2x2 over the whole slice was 85.9 and 71.2, with and without the overdiscrete B0 correction, respectively.
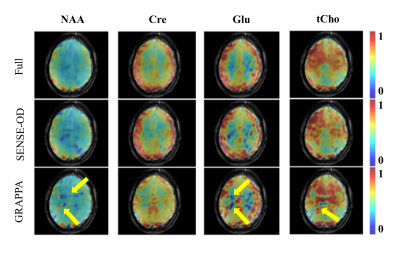
Judging by the previous results, the highest reliable acceleration factor was chosen to be R=2x2. Figure 4 shows the results of the OD-SENSE versus conventional GRAPPA reconstruction on the same volunteer. It can be seen that the OD-SENSE reconstruction performs superior to the conventional GRAPPA, as the conventional GRAPPA fails to resolve all of the aliasing and leaves traces of lipid rings in the maps.
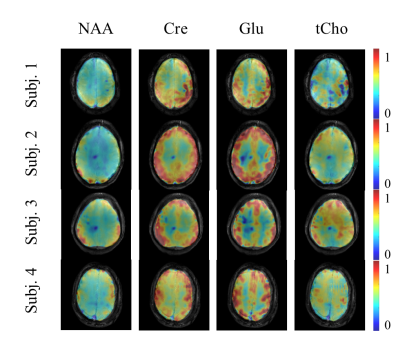
Figure 5 shows the reproducibility of the OD-SENSE reconstruction scheme for high resolution metabolite mapping (with R=2x2) on four healthy volunteers. As can be seen, high quality metabolite maps can be acquired in a reproducible manner.
Conclusion
In non-lipid suppressed MRSI, the lipid signals
in the subcutaneous region are orders of magnitude larger than the metabolite
signals. Therefore, any residual aliasing artifacts resulting from conventional
SENSE or GRAPPA reconstruction will overshadow the metabolites in the brain.
The over-discrete reconstruction scheme greatly reduced this artifact. The
over-discrete B0 correction resulted in 1.2 times gain in SNR. This
reconstruction scheme enabled high resolution metabolite mapping in about 3.5
minutes.Acknowledgements
No acknowledgement found.References
[1] Henning et al, NMR in Biomed 2009
[2] Nassirpour et al, Neuroimage 2017
[3] Bogner et al, MRM 2012
[4] Boer et al, MRM 2012
[5] Kirchner et al, MRM 2014
[6] Kirchner et al, MRM 2016
[7] Uecker et al, MRM 2014
[8] Blaimer et al, MRM 2006
Figures