3831
Detecting Glioblastoma invasion using Multi-parametric MRI and Quantitative Assessment with in-plane Histology1Glasgow Experimental MRI center, University of Glasgow, Glasgow, United Kingdom, 2Institute of Cancer Sciences, University of Glasgow, Glasgow, United Kingdom, 3MRI and clinical physics, Glasgow, United Kingdom
Synopsis
We evaluate the ability of a range of MRI techniques to probe glioblastoma invasion in a mouse model by comparison with in-plane stacked histology, enabling a direct voxel-to-voxel comparison between MRI and histology (HLA stain). We used the G7 mouse model which exhibits highly invasive tumour margins and scanned using T1 weighted, T2 weighted, T2map, Diffusion Tensor Imagining (DTI) and Perfusion Weighting Imaging (PWI). Registration of MRI datasets with stacked in-plane histology allows direct quantitative validation using DICE and ROC analysis, showing that PWI gave the best indication of tumour cell invasion.
Introduction
Glioblastoma multiforme (GBM) is the most common and aggressive primary brain tumour with poor prognosis with an average survival of 6-12 months after diagnosis with available treatments (1). A major difficulty to successful treatment is the ability of GBM cells to invade healthy brain tissue, making complete removal of a tumour by surgery difficult. Several studies have shown that tumour cells extend beyond borders defined by conventional Magnetic Resonance Imaging (cMRI) (2). In this study, we evaluate the ability of a range of MRI techniques to probe glioblastoma invasion in a mouse model by comparison with in-plane stacked histology, enabling a voxel-to-voxel comparison between MRI and histology.Methods
The experiment was performed with the G7 Glioblastoma mouse model, which derives from a primary human tumour cell line. All CD1 nude mice (n=10, 20-25g) were implanted intracranial with (5x105 cells per mouse) which is rich in tumour stem cells and highly invasive at the margins (3). MR imaging was performed on a Bruker 7T Biospec scanner with 72 cm volume resonator birdcage coil and a 4-channel phased array surface coil. Several imaging modalities were acquired as follows: T1w, T2w, DWI, ADC, and perfusion weighted multi boli Arterial Spin Imaging (mbASL) (4).
Histology slices were carefully cut to exactly match the orientation of the MRI slice and stained by Human Antigen Leukocyte (HLA) and H&E which allows probing tumour G7 cancer cells.To recreate the MRI slice thickness (1.5mm), three histology slices (20 m) from the front, middle, end of MRI slice were selected and averaged. The histology slices were coregistered linearly with MR images by using the Mutual Information method (5) .This allowed quantitative evaluation of the MRI scans as a biomarker of GBM invasion.Post processing of the data (with in-house developed MATLAB code) leads to a 3D matrix of the same dimensions and resolution. Following these steps, the abnormal region on each of image modality was manually selected in order to perform MRI-histology comparison tests (Dice, Sensitivity, Specificity and Accuracy).
Results
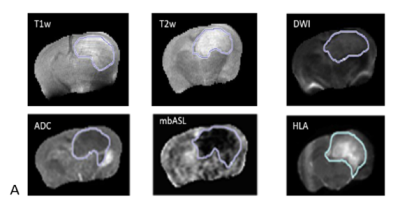
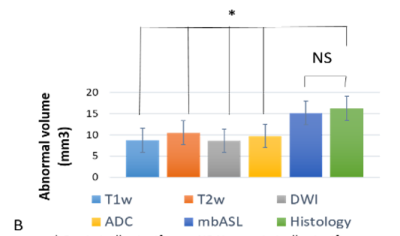
The histology abnormal region of interest (tumour) is overlaid to MRI data.Figure-1- shows several MRI modality and in-plane tumour maps from the same plane and resolution. Tumour volume is selected manually for histology slice and compared with cMRI tumour volume. cMRI probe significantly lower tumour volume than perfusion weighted map obtained by mbASL and histology (see Figure-2-).
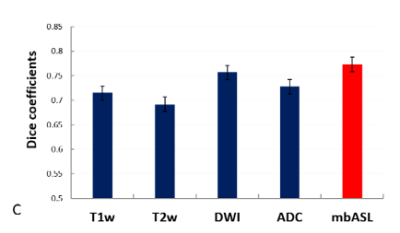
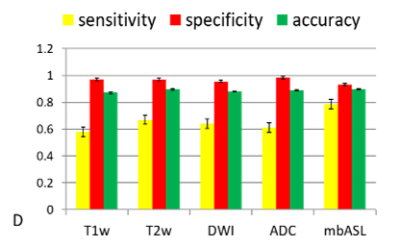
ROIs of abnormal regions were selected for each imaging modality and compare with histology. The statistical tests (Dsc, accuracy, specificity and sensitivity) are calculated and shown in Figure -3,4-.All imaging modality were showing high indexes with mbASL higher dice and sensitivity and lower specificity.