3768
MR Spectroscopy Biomarker Discovery Algorithms (MBDA) to Discriminate PTSD from Blast Exposure and Healthy Controls1Translational Research Institute, Brisbane, Australia, 2Queensland University of Technology, Brisbane, Australia, 3Draper, Boston, MA, United States, 4University of Newcastle, Newcastle, Australia
Synopsis
Single Voxel Point RESolved Spectroscopy (PRESS) was used to obtain neuro-metabolite and lipid levels from the Posterior Cingulate Gyrus (PCG) of 4 cohorts – Control, Civilian PTSD, Artillery and Close Combat soldiers. Using a wavelet based statistical analysis approach classifiers were developed to objectively diagnose and monitor neuro-deregulation associated with PTSD and blast injury to artillery and to close combat soldiers.
INTRODUCTION:
Despite the high prevalence of PTSD in frontline defenders (7%)[1-3] no objective diagnosis or biomarker of this disease process has been developed. Current diagnosis is based on self-report of symptoms[4]. Nor is there an objective test to evaluate the effect of blast exposure in front-line defenders. We reported previously that in vivo neuro two-dimensional COrrelated SpectroscopY could distinguish between these categories using conventional evaluation methods [5]. The size of the differences were large. The aim is to investigate the one dimensional MRS results with a wavelet-based algorithm post-processing approach and develop classifiers to use as biomarkers for PTSD and blast exposure.METHODS:
Data Acquisition: There were four participant groups – Healthy Control (N=33), PTSD (N=26) (civilian), Artillery (N=13) and Close Combat (N=8). These cohorts were screened by psychologists and determined to have only one condition. The controls were evaluated and had no neurological issues. The Artillery and Close Combat groups were exposed to percussive blast with the Close Combat (Breacher) group exposed to a much higher level of blast. All data were collected on a Siemens PRISMA scanner with a 64 channel head and neck coil. A 3D MPRAGE (TR/TE=2530/1.7 ms, 12-degree flip angle, FOV= 256x256mm, voxel size 1x1x1mm, NEX 4) was used for voxel localisation. MRS data were obtained using Point RESolved Spectroscopy (PRESS) (TE/TR 33/1500 ms) with a 3x3x3cm voxel placed in the Posterior Cingulate Gyrus (PCG).
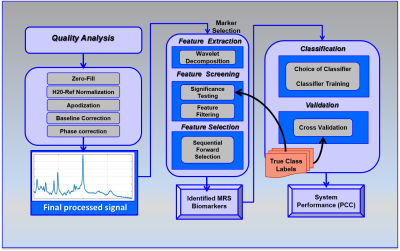
Data Processing: A new approach was used to analyse MRS signals to extract a rich set of wavelet-based features to enable development of a statistical classifier [6]. By evaluating each data point in the 4096 collected, the wavelet-based method allows discovery of previously unknown signatures not observed by traditional methods of analysis. The post-acquisition processing consists of a series of algorithms operating on the raw MRS signals to correct signal quality issues, remove the residual water signal, perform phase correction and baseline removal and compute the wavelet decomposition to represent features embedded in the signal (Figure 1). Feature extraction was performed on the real-valued absorption spectra derived from post-processed MRS spectra using wavelets [7] to extract features that are local to specific intervals in the frequency domain. Features were evaluated using a combination of statistical criteria to identify those statistically significant wavelet-based features that are least likely to be attributable to random effects (noise). This approach isolates just those features that discriminate between the two groups but also maximizes the probability that the features are due to distinct biochemical differences between the two groups. In each binary comparison, several wavelet-based features that met the thresholding criteria were identified.
To demonstrate classification performance, we performed 100 iterations of 5-fold cross-validation of a Linear Discriminant Analysis classification method, using a Sequential Forward Selection (SFS) scheme to identify optimal subsets of features to discriminate between classes. Because the study was conducted across multiple sites using different scanners, group differences could be confounded with the site/instrument effects. To clarify general linear modelling on the pooled data (Control, Civilian PTSD, Artillery, and Close Combat) was used to estimate the relative effects.
RESULTS:
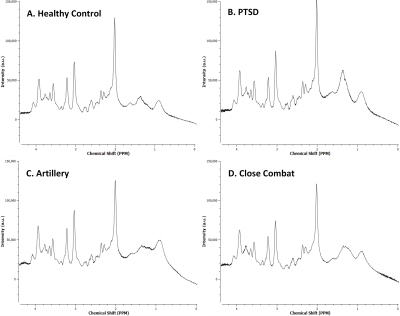
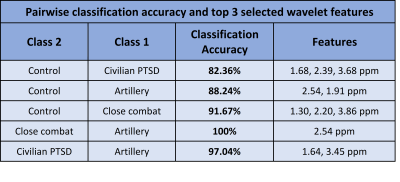
Typical 1D MR spectra from each cohort are shown in figure 2. There are noticeable but small differences. Figure 3 contains the average Percent Correct Classification from all iterations of the cross-validated SFS search for, at most, the top 3 features selected.
The highest accuracy was between the Close Combat cohort and the artillery with 100% accuracy; PTSD and Artillery 97%; Healthy control and Artillery 88%; and Healthy control and PTSD 83%. The classification of civilian PTSD and close combat cohort was however below 80%. There are clear categories of the clinically defined condition and these can be identified spectroscopically and are clear in the first three separations.
DISCUSSION:
Classifiers can be developed from the neuro 1D MRS of frontline defenders with blast exposure and PTSD using wavelet algorithm-based data processing. This has been shown to separate chronic pain from no pain [6]. There is a clear distinction between artillery soldiers exposed to blast and close combat soldiers.
This result is consistent with the evaluation of the 2D COSY where neuro-metabolite up-regulation was recorded for PTSD and head injury but down-regulation for blast exposure[5].
CONCLUSION:
Classifiers could be clinically applied to objectively diagnose and monitor neuro-deregulation associated with PTSD and blast injury. Our data suggest that a wavelet-based algorithm approach to utilising 1D MRS data may prove to be even more significant when combined with 2D L-COSY.Acknowledgements
Funding was provided by the US and Australian DoD. The authors acknowledge the support of radiographers Jameen Arm, Kylie Waters, Claire Berry and Antoinette Sweeney for implementing the protocols and running the scanner. We thank Defence personnel Drs Helen Cartledge, Patrick Cullinan and Amanda Toman for operational aspects.References
REFERENCES:
1. Sheerin, C.M., et al., The genetics and epigenetics of PTSD: overview, recent advances, and future directions. Current Opinion in Psychology, 2017. 14: p. 5-11.
2. Olff, M. and M. van Zuiden, Neuroendocrine and neuroimmune markers in PTSD: pre-, peri-and post-trauma glucocorticoid and inflammatory dysregulation. Current Opinion in Psychology, 2017.
3. Gilam, G. and T. Hendler, Tracing the Neural Carryover Effects of Anger and their Relation to Traumatic Stress Symptoms. Biological Psychiatry, 2017. 81(10): p. S165-S166.
4. Association, A.P., Diagnostic and statistical manual of mental disorders (DSM-5®). 2013: American Psychiatric Pub.
5. Galloway, G.J., et al., Neuro 2D correlated spectroscopy identifies neuro deregulation in soldiers exposed to blast prior to discernible changes By conventional imaging. in Int.Soc.Magn.Reson.Med. 2017. Hawaii., 2017.
6. Stanwell, P., et al., Neuro magnetic resonance spectroscopy using wavelet decomposition and statistical testing identifies biochemical changes in people with spinal cord injury and pain. Neuroimage, 2010. 53(2): p. 544-52.
7. Daubechies, I., Ten lectures on wavelets. 1992: SIAM.
Figures