3672
The effects of LIS1 deletion in the adult mouse brain and partial rescue by LiCl: A T2 mapping, magnetization transfer and diffusion MRI study1Department of Veterinary Resources, Weizmann Institute of Science, Rehovot, Israel, 2Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel
Synopsis
LIS1, a gene mutated in lissencephaly (“smooth brain”) have been investigated mainly in the developing brain. Initial studies demonstrated distinct ataxia and rapid lethality following Lis1 conditional deletion in adult mice. Therefore, our aim was to investigate the postnatal roles of LIS1 and the underlying mechanism. Conditional Lis1 knockout mice studied by MRI pre, and 5 days post, tamoxifen-induced Lis1 deletion, showed increase in T2 and ADC and decrease MTR and FA0, in the lateral ventricles and in brain regions related to motion and auditory functions. These alterations and changes in the Wnt pathway were partially rescued by LiCl treatment.
Introduction
LIS1 is a gene mutated in lissencephaly, a neuronal migration disorder in the developing brain (“smooth brain”). LIS1 is involved in regulation of the microtubule cytoskeleton and participates in several protein complexes and thus connected to multiple cellular activities such as cell migration, intracellular transport, cell polarity, mitosis and meiosis1. LIS1 activities have so far been investigated mainly in the developing brain2. Our initial studies demonstrated distinct ataxia and rapid lethality following Lis1 conditional deletion in adult mice that have not been documented before3. Thus, the aim of this MRI study was to investigate the postnatal roles of LIS1 and the underlying mechanism.Methods
Mouse model: Tamoxifen induced conditional Lis1 knockout (Lis1 cKO; 129S-Pafah1b1tm2Awb/J crossed with B6.Cg-Tg(UBC-cre/ERT2)1Ejb/1J) mice were studied before and at day 6, following daily tamoxifen administration (125 mg of tamoxifen in corn oil per kg of body weight, injected i.p for 5 consecutive days).
Drug Treatments: Lithium chloride (LiCl, sigma) was provided in drinking water (1.2 g/L; starting two weeks before tamoxifen administration). The control group was provided with regular drinking water.
MRI experiments: MR images were acquired on a 9.4T horizontal Bruker Biospec using a linear coil for excitation and a receive quadrature head surface coil for detection. T2 mapping: MSME, TR 3000 ms; 16 echoes, TE 10-160 ms; two averages; matrix 256 x 256; FOV 1.8 x 1.8 cm2; 15 slices of 1 mm w/o gap; acquisition time 12min48s. Magnetization transfer: RARE, RARE factor 6; TR 2500 ms; TE 27 ms; irradiation offset 2000 Hz; MT 0 and 25 μT; two averages; matrix 256 x 265; FOV 1.8 x 1.8 cm2; 15 slices of 1 mm w/o gap; acquisition time 1min45s. Diffusion: DTI-SE-EPI, TR 2000 ms; TE 23 ms; Δ 10 ms, δ 4.5 ms, b-value 650 s/mm2; 30 directions; no averaging; matrix 128 x 128; FOV 1.8 x 1.8 cm2; 11 slices of 1 mm w/o gap; acquisition time 4min8s. T2 and MTR maps were calculated using in-house MATLAB scripts. ADC and FA0 maps were generated using dsi-studio software. Co-registration inter-subject and intra-subject was applied before the MRI dataset analysis. For optimal suitability to a mouse brain atlas (correction of head movements image artifacts), all images went through atlas registration: reslicing, realignment and smoothing, using the SPM12 software. SPM12 software was also used to evaluate the statistical significance between the maps of pre and post induction of LIS1 deletion (paired-t-test, p<0.05) and the statistical significance of LiCl treatment (ANOVA, p<0.05).
Ex vivo studies: Mice brains were excised and processed for immunostaining and RT-PCR.
Results
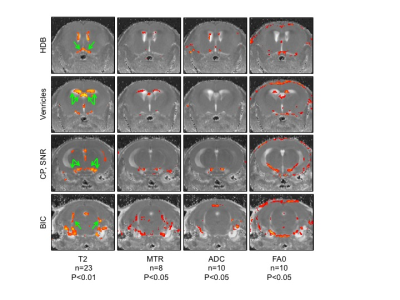
Lis1 cKO mice exhibited a severe ataxic phenotype that was clearly visible after three daily tamoxifen injections (induction of Lis1 deletion) and was lethal within 6-7 days. Initial MRI and histopathology suggested that the mice developed acute hydrocephalus. However, consecutive MRI experiment done on the same mice, before and on day 6 (following 5 daily tamoxifen injections), indicated that some of the mice had enlarged ventricles prior to treatment, suggesting the Lis1 floxed allele is a hypomorph. Importantly, the images pointed to specific brain areas that were significantly altered, showing increase in T2 and ADC and decrease in MTR and AF0 in response to tamoxifen injection (Figure1). The highlighted areas may be responsible for the observed phenotype and include the following regions: HDB, nucleus of the horizontal limb of the diagonal band, involved in spinocerebellar ataxia type 24. CP, cerebellar peduncle, involved in motor vestibular functions such as balance and posture maintenance5. SNR, substantia nigra (reticular part) has an important role in motion. BIC, brachium of the inferior colliculus, part of the auditory pathway.
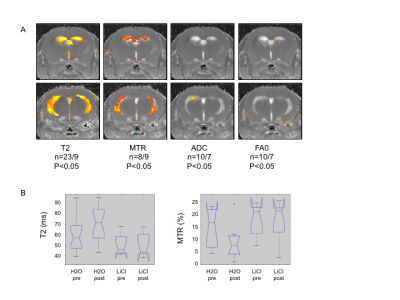
LIS1 mutant organoids exhibited changes in the Wnt pathway6, and we noted changes in the ventricular region of the Lis1 cKO mice by qPCR (significant differences in Wnt7b, cyclin D1, DVL1-3, mCatnb). These alterations were partially rescued by LiCl treatment. LiCl treatment also resulted in partial rescue in our Lis1 cKO mice, by improving the ataxic phenotype and MRI T2 and MTR parameters in the ventricles regions (Figure2), suggesting alleviation of edema and tissue destruction caused by Lis1 deletion.
Discussion and Conclusion
The decrease in MTR reflects decrease in macromolecule content and suggests destruction of myelin sheets, resulting in more isotropic water diffusion and therefore decrease in FA0 and increase in ADC. The increase in T2 on the other hand, may reflect edema, inflammation or destruction of membranes. Altogether, these findings contribute to our understanding of the specific role of LIS1 in the mature brain and the associated neuronal and signaling pathways.
Acknowledgements
No acknowledgement found.References
1. Reiner O, Sapir T. LIS1 functions in normal development and disease. Curr Opin Neurobiol. 2013;23(6):951-6.
2. Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41(2):168-77.
3. Hirotsune S, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19(4):333-9.
4. Rub U, et al. Involvement of the cholinergic basal forebrain nuclei in spinocerebellar ataxia type 2 (SCA2). Neuropathol Appl Neurobiol. 2013;39(6):634-43.
5. Cavallari M, et al. Mobility impairment is associated with reduced microstructural integrity of the inferior and superior cerebellar peduncles in elderly with no clinical signs of cerebellar dysfunction. Neuroimage Clin. 2013;2:332-40.
6. Iefremova V, et al. An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell reports. Cell Rep. 2017;19(1):50-59.
Figures