3528
Deep-SENSE: Learning Coil Sensitivity Functions for SENSE Reconstruction Using Deep Learning1Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Shenzhen, China, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Synopsis
Parallel imaging is an essential tool for accelerating image acquisition by exploiting the spatial encoding effects of RF receiver coil sensitivity functions. In practice, the coil sensitivity functions are often estimated from low-resolution auto-calibration signals (ACS) which limits estimation accuracy and in turn results in aliasing artifacts in the final reconstructions. This paper presents a novel deep learning based method for coil sensitivity estimation which exploits empirical and physics-based prior information to produce high-accuracy estimates of coil sensitivity functions from low-resolution ACS. Results are given which demonstrate the proposed method provides a significant reduction in aliasing over standard methods.
Introduction
Parallel imaging accelerates image acquisition by exploiting the spatial encoding effects of the RF receiver coil sensitivity function1. In practice, the coil sensitivities are often estimated from auto-calibration signals (ACS) and their accuracy is usually limited by the amount of ACS data measured. Poor estimates of the coil sensitivity functions can lead to image reconstruction errors in the form of aliasing artifacts. This paper presents a novel machine learning based method for the estimation of coil sensitivity functions. The proposed method exploits the fact that for a given receiver system, the coil sensitivity functions (as a function of both coil geometry and subject loading effects) can be first measured experimentally under various experimental conditions to create a set of “a priori” coil sensitivity functions. These a priori sensitivity functions can then be used to train a deep neural network to predict/estimate the coil sensitivity functions for a specific experiment using limited ACS data.Methods
The proposed method is illustrated in Figure 1. We construct a multi-level deep learning approach to integrate empirical and physics-based prior information to produce high-accuracy sensitivity estimates. To incorporate the prior knowledge of coil sensitivity functions $$$Z$$$ obtained from “training” scans, a deep neural network (Net1) is designed. This network was trained to generate high-resolution sensitivity maps $$$Y_1$$$ given both $$$Z$$$ and $$$X$$$, where $$$X$$$ is a coil sensitivity estimated from limited ACS lines. The coil sensitivity functions thus generated can be viewed as a “best” estimate based on the functional relationships between the low-resolution and the high-resolution versions of a coil sensitivity function captured in the prior sensitivity functions as well as the ACS data newly acquired from a specific experiment.
To further improve the estimates of the coil sensitivity functions with limited ACS data, the proposed method uses a second neural network (Net2, e.g., auto-encoder) to incorporate (physics-based) prior knowledge of coil sensitivity functions. The second neural network exploits the facts that under certain mild conditions, the sensitivity functions can be represented by a parametric model. Such a physics-based model implicitly uses a subspace of features to represent the desired coil sensitivity profiles. Learning is then implemented in this subspace, which is expected to perform better due to a reduction in the degree of freedom especially when there is only limited training data. More specifically, in this work, we employ a 3D polynomial model and learn the mapping between the polynomial coefficients of the low-resolution and the high-resolution versions of the sensitivity maps.
To address any potential problem with inaccurate predictions from Net1 and Net2, a third neural network (Net3) is proposed to integrate $$$Y_1$$$ and $$$Y_2$$$ to produce the final desired sensitivity function $$$Y$$$. $$$Y_1$$$ includes and utilizes both prior sensitivity knowledge and subject dependent variations (i.e., subject loading and position) while $$$Y_2$$$ benefits from the use of physical constraints. The effective use of the complementary information in $$$Y_1$$$ and $$$Y_2$$$ is a key to our method’s success to predict coil sensitivity functions.
Materials
Data were acquired on a 3T MR scanner (SIEMENS Prisma) from 5 subjects using 3D TSE and GRE sequences (matrix size 224$$$\times$$$224, FOV 240mm$$$\times$$$240mm, slice thickness 2mm, FA 90°. For GRE, TR/TE 20/2.24ms, while for TSE, TR/TE 3200/220ms. $$$X$$$ used for training were derived from 28 central ACS lines.
For Net1 and Net3, we employed a convolutional neural network known as U-net2 which was implemented in Tensorflow 1.3.0 and trained using stochastic gradient descent (SGD) with an Adam optimizer. The magnitude and phase maps were trained separately. Value augmentation (e.g., random flipping and rotation) were implemented to provide more datasets for training. SENSE3 reconstructions were computed using both our learned sensitivity maps and low-resolution sensitivity maps with uniform undersampling.
Results and Discussion
Figure 2 shows the magnitude and phase components of the sensitivities estimated from the ACS lines using the adaptive combined method4 and the predicted ones by our proposed method, respectively. As can be seen, the sensitivity functions from our method exhibit higher quality, and lead to better SENSE reconstruction as shown in Fig. 3. Note that reconstructions made using our sensitivity contain much less aliasing artifacts across all the reduction factors tested.Conclusion
This work proposes a novel deep learning method to learn coil sensitivity functions for SENSE reconstruction. The new method produces significantly improved coil sensitivity functions compared to those obtained directly from the limited ACS lines, thus leading to improved SENSE reconstructions at high reduction factors.Acknowledgements
This work was partially supported by NIH-R21-EB021013-01, NIH-P41-EB002034 and NSFC-61671441.References
- Ying L et al. Parallel MRI using phased array coils. IEEE SignalProcess Mag 2010;27:90–98.
- Ronneberger O et al. U-net: Convolutional networks for biomedical image segmentation, MICCAI, 2015;234-241.
- Pruessmann KP et al. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952–962.
- Walsh DO et al. Adaptive reconstruction ofphased array MR imagery. Magn Reson Med 2000;43:682–690.
Figures
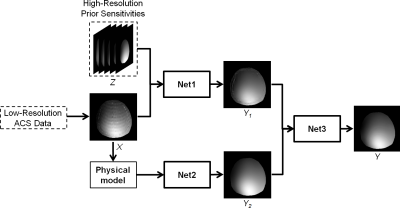
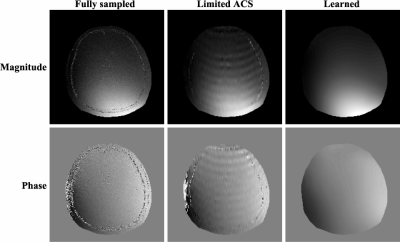
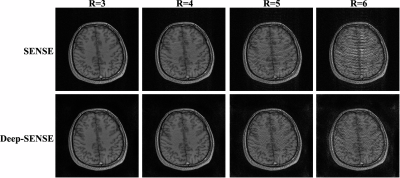
Figure 3. SENSE reconstructions using sensitivities directly estimated from limited ACS data (top row) and from the proposed method (bottom row). The Deep-SENSE reconstructions contain much less aliasing across all the reduction factors tested.