3525
MR Fingerprinting using a Gadgetron-based reconstructionWei-Ching Lo1, Yun Jiang2, Dominique Franson1, Mark Griswold1,2, Vikas Gulani1,2, and Nicole Seiberlich1,2
1Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Department of Radiology, University Hospitals Cleveland Medical Center at Case Western Reserve University, Cleveland, OH, United States
Synopsis
Gadgetron-based online MRF reconstruction enables rapid generation of quantitative tissue property maps directly at the scanner before completing acquisition of the following slice. This technique can facilitate multicenter clinical studies and facilitate easier and direct comparisons of quantitative maps from different scanners.
Purpose
Magnetic Resonance Fingerprinting (MRF) (1,2) is a recently developed MR technique that allows simultaneous generation of quantitative maps of multiple tissue properties. MRF was originally implemented in Matlab, such that all processing was performed off-line and involved cumbersome data transfer, making clinical translation challenging. Here, an online reconstruction is demonstrated, using a Gadgetron-based framework to enable rapid generation of MRF derived tissue property maps directly at the scanner. Furthermore, online MRF reconstruction may facilitate multicenter clinical studies and facilitate comparison of quantitative maps from different scanners.Methods
Experiments were performed on a 3T Skyra scanner (Siemens Medical Solutions, Erlangen, Germany) using the MRF-FISP acquisition (2) with the following parameters: FOV = 400x400 mm2; matrix size = 400x400; in-plane resolution = 1x1 mm2; flip angle = 5-75°; TR = 12-15 ms; slice thickness = 5 mm. The acquisition time for a single 2D slice was 45 seconds. The MRF dictionary was generated using Bloch equation simulations in MATLAB (MathWorks 2015b, Natick, MA) with T1 resolution of [10:10:100 120:20:1000 1040:40:2000 2050:150:2950 3100:100:4500] and T2 resolution of [2:2:100 105:5:150 160:10:300 350:50:800 900:100:1600 1800:200:3000], denoted by min:step:max (ms). The dictionary included a total of 8,537 entries. The computer used to perform online Gadgetron reconstructions has an 8GB Nvidia GeForce GTX 1080 graphics card; a 10 core, 2.2GHz Intel Xeon E5-2630 v4 processor; and 64GB of 2400MHz DDR4 RAM. During MRF acquisition, the PCA-based coil compressed raw data (3) were accumulated and passed on to the Gadgetron reconstruction pipeline along with the pre-calculated dictionary. To further reduce the computational load and memory requirements without reducing performance, SVD basis compression (4) was applied to the MRF data to compress the number of time points from 3000 to 52. The SVD compressed data were then gridded using GPU-enabled NUFFT (5) and combined via adaptive coil combination (6). Direct pattern matching was applied to the data to extract quantitative T1, T2, and proton density maps. All Gadgetron-based quantitative maps were passed back to the scanner to generate DICOM images for further post-processing. The accuracy of the reconstruction was validated using the ISMRM/NIST MRI system phantom (7). The mean and standard deviation of each compartment were calculated from 70 pixels within a circular ROI that was manually drawn on the M0 map. The results from the proposed online Gadgetron framework and from an offline MATLAB reconstruction were compared to the gold standard IR-SE method for T1 values and the multiple single-echo spin echo method for T2 values (7). The MATLAB reconstruction followed all of the steps in the Gadgetron framework, but no coil compression was used.Results
The total reconstruction time for the online Gadgetron framework was approximately 20 seconds per slice, while the total reconstruction time for the offline reconstruction using MATLAB was approximately 190 seconds per slice. The time required to perform the most time-consuming steps in the reconstruction process are shown in Table 1. Figure 1 shows mean T1 (a) and T2 (b) values from MRF with Gadgetron and MRF with MATLAB reconstruction plotted against the values from the gold standard reference for each phantom compartment and each of the three measurements. The results from the Gadgetron framework are in excellent agreement with the MATLAB reconstruction; the greatest absolute percentage errors between the Gadgetron framework and the MATLAB reconstruction were less than 0.5% for T1 and 0.9% for T2. Figure 2 showed demonstrative T1 and T2 maps for brain and prostate generated using the Gadgetron framework. The error mean, error standard deviation (SD) and structural similarity index (SSIM) for brain and prostate (Table 2) demonstrates that quantitative maps can be generated using the Gadgetron framework with negligible errors.Discussion
This Gadgetron-based online MRF reconstruction can be used to generate quantitative maps rapidly at the scanner before completing acquisition of the following slice. Despite the additional coil compression step in the Gadgetron reconstruction, the T1 and T2 values as measured in the ISMRM/NIST phantom were in excellent agreement with those generated using the standard MATLAB reconstruction. This framework could enable clinical translation of MRF and employment of the technology in a clinical setting. Furthermore, the Gadgetron-based MRF reconstruction is applicable to different commercial MR scanners for large scale multicenter and multivendor studies. As an example, the MRF Gadgetron reconstruction has been deployed on seven workstations in three different medical institutions, further demonstrating the advantages of a scanner-agnostic reconstruction.Conclusion
This work enables rapid online reconstructions for MRF tissue property mapping using a Gadgetron-based framework with coil compression, singular value decomposition, and direct pattern matching.Acknowledgements
Siemens Healthineers, 1R01EB016728, 1R01DK098503, 1R01CA208236. Thanks to Hua Wei, Hui Xue and Kelvin Chow for their contributions to this work.References
[1] Ma et al. Nature. 2013;495:187–192. [2] Jiang et al. MRM. 2015;74:1621–1631. [3] Hansen et al. MRM. 2013;69:1768–1776. [4] McGivney et al. IEEE TMI. 2014;33:2311–2322. [5] Sorensen et al. IEEE TMI. 2008;27:538–547. [6] Inati et al. Proc. of ISMRM. 2013, #2672. [7] Jiang et al. MRM. 2016.Figures
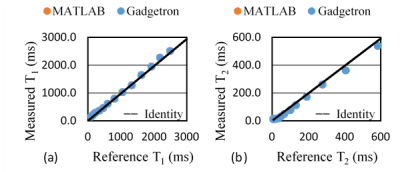
Figure 1. The mean T1 (a) and T2 (b) values from
MRF with Gadgetron (blue) and MRF with MATLAB (orange)
plotted
against the gold standard reference values. Note that the Gadgetron and MATLAB values lie directly on top of one
another.
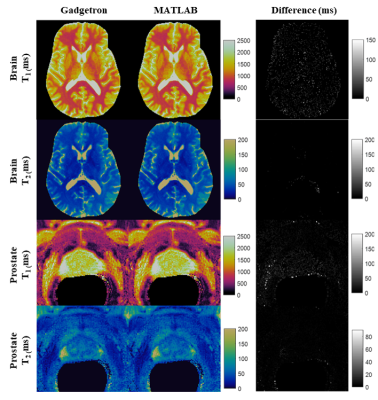
Figure 2. Demonstrative T1 and T2 maps (cropped) for brain and
prostate generated using the Gadgetron framework and MATLAB
reconstruction.
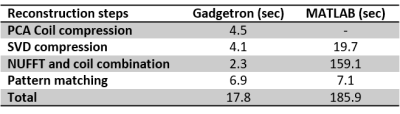
Table 1. Time required for different steps in the
reconstruction process. Reported times are averaged over 3 reconstructions of
the same dataset.
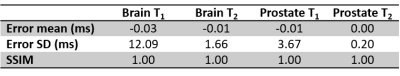
Table 2. Error mean, error SD, and SSIM between MATLAB and Gadgetron in brain and
prostate.