3523
A Python-based MRI Reconstruction Toolbox, “MRIPY”, for Compressed Sensing, Parallel Imaging and Machine Learning1Department of Radiology and Biomedical Imaging, University of California at San Francisco, San Francisco, CA, United States
Synopsis
A python-based open-source package, “MRIPY” combines the existing MRI reconstruction methods, i.e. compressed sensing and parallel imaging, with deep neural networks that are implemented in the Tensorflow software.
Purpose
The purpose of this research project is to develop and validate an open-source toolbox for next-generation accelerated MRI reconstruction. This abstract presents a python-based open-source package as the output of this project, developed to combine the existing MRI reconstruction methods, i.e. compressed sensing and parallel imaging, with deep neural networks that can be integrated with software such as Tensorflow and PyTorch.Methods
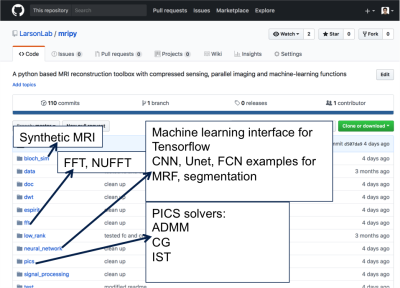
Fig. 1 shows a few key functions of this python package (https://github.com/LarsonLab/mripy/):
- MRI reconstruction with compressed sensing 1 via convex optimization algorithms: gradient descent, nonlinear conjugate gradient and alternating direction method of multipliers 2,3
- Parallel imaging, ESPiRIT 4, which is a Hankel low rank and PCA based analysis method in combination with compressed sensing reconstruction 4
- Interface with Tensorflow (https://www.tensorflow.org) for training and testing neural networks in classes/functions that can be easily utilized in the abovementioned optimization algorithms during MRI reconstruction/processing
- FFTW (https://hgomersall.github.io/pyFFTW/) /cuFFT (https://developer.nvidia.com/cufft) /DWT (https://pywavelets.readthedocs.io/en/latest/) interfaces, and custom-built 1D/2D/3D non-uniform FFT 5 with optional GPU acceleration capabilities (implemented in CUDA and numba)
- Numerical MRI signal simulation with optional GPU acceleration capabilities (implemented in CUDA and numba)
Results
Figures 2-5 show sample outputs generated from MRIPY, including a range of MRI reconstruction methods as well as integration with deep neural networks:
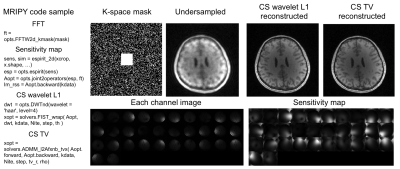
Fig. 2 shows an example of parallel imaging and compressed sensing reconstruction with alternating direction method of multipliers (ADMM) and wavelet L1 or total variation minimization regularization. Raw MRI data was from website (http://people.eecs.berkeley.edu/~mlustig/CS.html).
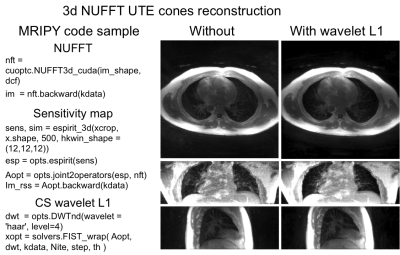
Fig. 3 shows NUFFT based reconstruction for UTE images (after MIP) acquired using a 3D UTE Cones sequence.
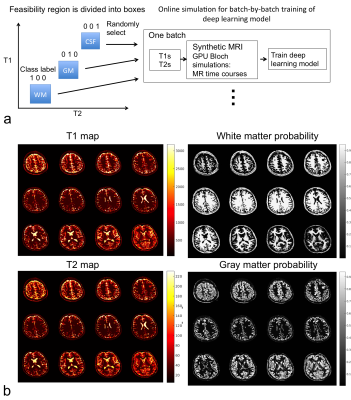
Fig. 4 shows whole brain segmentation/relaxometry as the direct output of a deep neural network trained with the synthetic MRI data. The in vivo brain MRI data was from Dr. Jing Liu.
Fig. 5 shows another more complex task that is the detection/segmentation of myocardium from MRI image. A fully convolutional network performed the segmentation as an example provide in MRIPY. The MRI data was from Dr. Yan Wang.
Conclusion
The new reconstruction and neural network segmentation methods developed will provide improved precise measurements in MRI.Acknowledgements
We receive grant support (NIH R21NS089004 and NIH R56HL133663 ) from NIH.References
1. Lustig, M. & Pauly, J. M. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn. Reson. Med. 64, 457–471 (2010).
2. Afonso, M. V., Bioucas-Dias, J. M. & Figueiredo, M. A. T. An augmented lagrangian approach to the constrained optimization formulation of imaging inverse problems. IEEE Trans. Image Process. 20, 681–695 (2011).
3. Boyd, S., Parikh, N., E Chu, B. P. & Eckstein, J. Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers. Found. Trends Mach. Learn. 3, 1–122 (2011).
4. Uecker, M. et al. ESPIRiT — An Eigenvalue Approach to Autocalibrating Parallel MRI : Where SENSE Meets GRAPPA. 0, 1–12 (2013).
5. Greengard, L. & Lee, J. Accelerating the Nonuniform Fast Fourier Transform ∗. 46, 443–454 (2004).
Figures