3522
Easy-to-Implement and Rapid Image Reconstruction of Accelerated Cine and 4D Flow MRI Using TensorFlow1Institute for Biomedical Engineering, ETH Zurich, Zurich, Switzerland
Synopsis
Many MR image reconstruction algorithms can be formulated as optimization problems and solved with gradient-based optimization methods of choice. In this work, we present and analyze the performance of the TensorFlow framework for modeling and solving MR image reconstruction problems. We test our approach on undersampled cine cardiac and 4D flow datasets. It is demonstrated that MR image reconstruction is easy to implement in TensorFlow, TensorFlow performs comparably to sophisticated optimization algorithms with theoretical convergence guarantees, and that TensorFlow is as fast as or faster compared to standard MR reconstruction toolboxes.
Introduction
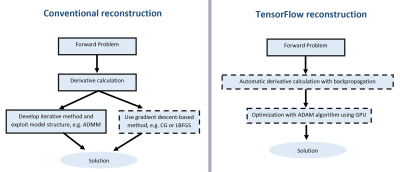
Modern MR image reconstruction algorithms are based on the optimization framework. In many applications, a target cost function is constructed using data fidelity and regularization terms which is then optimized using a gradient-based method of choice, such as IRLS [1] or the alternating direction method of multipliers (ADMM) [2,3] among many others. For their efficient implementation, it is essential to calculate gradients of the cost function, which is usually conducted manually. In this work, we present and analyze the performance of the TensorFlow framework [4] applied for MR image reconstruction. TensorFlow allows defining a computational graph that implements an acquisition model using a convenient Numpy-like set of operations. The gradient of the cost function with respect to optimizable signal properties is then inferred automatically using the backpropagation algorithm [5]. The optimization is performed with the ADAM algorithm [6], automatically parallelizing computations using GPU. The approach is tested on undersampled cardiac cine and 4D flow datasets [7] and compared to reconstructions using the BART [2] in terms of accuracy and speed.Methods
Cine MRI
Fully sampled short-axis cardiac cine datasets were acquired on 3T system (Philips Healthcare, Best, The Netherlands) with spatial resolution of 1.4x1.4 mm3 and $$$N_t=25$$$ cardiac phases. Variable density and temporally incoherent 5-fold undersampling pattern was simulated for each frame. Applying the Fourier transform in the temporal domain to sparsify the signal, the reconstruction problem reads:
$$\min_{X\in\mathbb{C}^{N_p\times\!\,N_t}}\|M\circ(F_pX-Y)\|_{2,2}^2+\lambda\|F_tX^T\|_{1,1}=\min_{X\in\mathbb{C}^{N_p\times\,N_t}}\mathcal{F}(X),\qquad\qquad(1)$$where $$$X$$$ is the image series estimate, $$$M\in\{0,1\}^{N_p\times\,N_t}$$$ is the undersampling mask, $$$F_p$$$ and $$$F_t$$$ are discrete Fourier transforms in spatial and temporal dimensions correspondingly, $$$Y$$$ is a matrix of zero-filled k-space samples, and regularization parameter $$$\lambda=0.004$$$ is determined empirically. ADMM [3] solution of the problem (1) is used as the baseline.To solve problem (1) in TensorFlow, we defined the corresponding computational graph as specified by the cost function. Since gradients of operations are provided by TensorFlow, the backpropagation algorithm calculates $$$\frac{\partial\mathcal{F}}{\partial\,\!X}$$$ via the chain rule. ADAM [6] algorithm is used to minimize $$$\mathcal{F}(X)$$$, with a step length of 0.01.
4D Flow MRI
Fully sampled 4D flow data in the aortic arch of a healthy volunteer were acquired on a 3T Philips Ingenia system (Philips Healthcare, Best, the Netherlands) using a navigated Cartesian four-point phase-contrast gradient-echo sequence with uniform venc of 200 cm/s and a spatial resolution of 1.83x1.83x1.83 mm3. The data were retrospectively undersampled to simulate an accelerated acquisition with factors ranging from 6 to 20. Coil sensitivity maps were calibrated with ESPIRiT [8], k-space data were compressed from 28 to $$$N_b=8$$$ virtual receiver channels [10]. Reconstruction was implemented using temporal total variation [9] regularization:
$$\min_{X_v\in\mathbb{C}^{N_p\times\,\!N_t}}\sum_{j=1,\dots,N_b}\|M_{j,v}\circ(W_{b_j}F_pX_v−Y_{j,v})\|_{2,2}^2+\lambda\|D_tX_v^{T}\|_{1,1},\quad\,v=1,\dots,N_v.\qquad(2)$$
Here $$$X_v$$$ is the estimate for $$$v$$$-th velocity encoding, $$$W_{b_j}$$$ is the diagonal matrix defined by the corresponding coil sensitivity map, $$$Y_{j,v}$$$ are zero-filled k-space samples, $$$M_{j,v}$$$ are the undersampling masks, $$$D_t\in\{−1,1\}^{N_t\times\,\!N_t}$$$ is the temporal finite difference matrix. In BART [2] problem (2) is solved using the ADMM algorithm employing the conjugate gradient method for internal iterations. In TensorFlow, the computational graph for problem (2) was defined in a similar way as described for problem (1) and optimized with the ADAM algorithm using 300 iterations.
Results
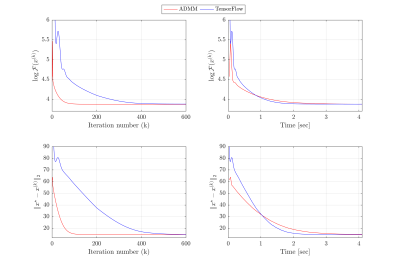
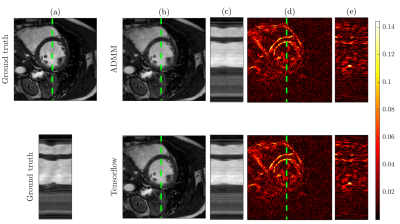
Figure 2 shows that both ADMM and TensorFlow reconstructions of the cine MRI data achieve the same performance in terms of optimization cost and estimated residual relative to ground truth. TensorFlow requires more iterations than ADMM to converge (378 for TensorFlow vs. 71 for ADMM). However, when iteration time is taken into account, due to GPU utilization, TensorFlow takes 1.6 seconds to converge, compared to 2.4 seconds using ADMM. Figure 3 illustrates that both reconstruction methods provide visually comparable results.
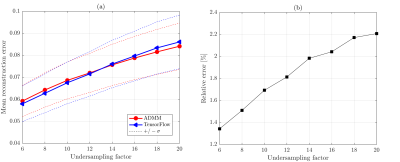
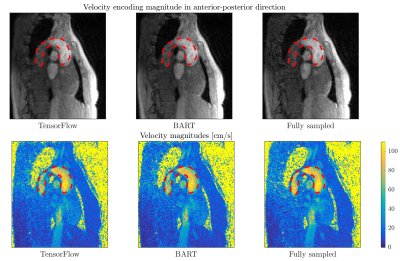
Figure 4 shows that both methods provide similar reconstruction accuracy for all acceleration factors tested on the 4D flow data. Relative velocity magnitude discrepancy between BART and TensorFlow reconstructions inside the aorta was less than 3%. Reconstruction results are illustrated in Figure 5. Average runtime of BART was 7.5 minutes on a 6-core 3.8 GHz CPU, runtime of TensorFlow was 4.6 minutes on NVIDIA Titan Xp GPU.
Discussion
Acknowledgements
The authors acknowledge funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 668039.
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research.
References
[1] Jung H, Sung K, Nayak KS, Kim EY, Ye JC. k‐t FOCUSS: A general compressed sensing framework for high resolution dynamic MRI. Magnetic Resonance in Medicine. 2009. 61(1):103-16.
[2] Uecker M, Ong F, Tamir JI, Bahri D, Virtue P, Cheng JY, Zhang T, Lustig M. Berkeley advanced reconstruction toolbox. In Proc. Intl. Soc. Mag. Reson. Med. 2015 (Vol. 23, p. 2486).
[3] Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Foundations and Trends in Machine Learning. 2011. 3(1):1-22.
[4] Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. 2016. arXiv:1603.04467.
[5] Griewank A, Walther A. Evaluating derivatives: principles and techniques of algorithmic differentiation. Society for Industrial and Applied Mathematics. 2008.
[6] Kingma DP, Ba JL: Adam: A method for stochastic optimization. In Proc ICLR. 2013. 1–15.
[7] Valvano G, Martini N, Huber A, Santelli C, Binter C, Chiappino D, Landini L, Kozerke S. Accelerating 4d flow Mri by exploiting low‐rank matrix structure and hadamard sparsity. Magnetic resonance in medicine. 2017 Oct 1;78(4):1330-41.
[8] Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magnetic resonance in medicine. 2014. 71(3):990-1001.
[9] Blomgren P, Chan TF. Color TV: total variation methods for restoration of vector-valued images. IEEE Transactions on Image Processing. 1998. 7(3):304-9.
[10] Buehrer M, Boesiger P, Kozerke S. Virtual body coil calibration for phased-array imaging. Proceedings of the 17th Annual Meeting of the ISMRM. 2009. (p. 3209).
Figures