3502
Pelvic Organ Segmentation with Sample Attention based Stochastic Connection Networks1Department of Computer Science, UNC-Chapel Hill, Chapel Hill, NC, United States, 2Department of Radiology and BRIC, UNC-Chapel Hill, Chapel Hill, NC, United States, 3Department of Radiation Oncology, UNC-Chapel Hill, Chapel Hill, NC, United States
Synopsis
Accurate segmentation of pelvic organs is important for prostate radiation therapy. Modern radiation therapy starts to use magnetic resonance image (MRI) as an alternative to CT image, because of the superior soft tissue contrast of MRI and also no risk of radiation exposure. In this abstract, we propose a novel deep network architecture, called “Sample Attention based Stochastic Connection Networks” (SASCNet), to delineate pelvic organs from MRI in an end-to-end fashion. Our proposed network has two main contributions: 1) We propose a novel randomized connection module and adopt it as a basic unit to combine the shallower and deeper layers in the fully convolutional networks (FCN); 2) We propose a novel adversarial attention mechanism to automatically dispatch sample importance so that we can avoid the domination of easy samples in training the network. Experimental results show that our SASCNet achieves competitive segmentation accuracy.
Purpose
Accurate segmentation of pelvic organs is important for prostate radiation therapy. Modern radiation therapy starts to use magnetic resonance image (MRI) as an alternative to CT image, because of the superior soft tissue contrast of MRI and also no risk of radiation exposure. In this abstract, we propose a novel deep network architecture, called “Sample Attention based Stochastic Connection Networks” (SASCNet), to delineate pelvic organs from MRI in an end-to-end fashion. Our proposed network has two main contributions: 1) We propose a novel randomized connection module and adopt it as a basic unit to combine the shallower and deeper layers in the fully convolutional networks (FCN); 2) We propose a novel adversarial attention mechanism to automatically dispatch sample importance so that we can avoid the domination of easy samples in training the network. Experimental results show that our SASCNet achieves competitive segmentation accuracy.Method
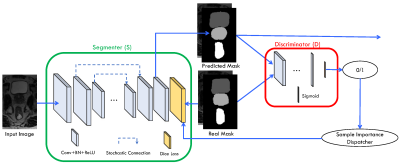
FCN1 has been widely adopted in various semantic segmentation tasks and achieved superior performance. While being successful, FCN cannot accurately localize object boundaries due to the lack of fine-level information during the label inference stage. To tackle this problem, Unet2 was proposed to combine low-level feature maps with the high-level feature maps together for label inference during the condensing process. This combination effectively addresses the limitation of FCN and also improves the localization accuracy. However, it could suffer from serious overfitting issues due to small dataset in medical image dataset. To overcome the problem, we propose to inject stochastic connection, instead of full connection, to combine the low-level and high-level feature maps (as shown in Fig. 1). Benefiting from the ensemble essence of the stochastic connection 3, the proposed segmentation network can alleviate the overfitting issues to a large extent.
Inspired by the generative adversarial networks 4, we design our segmentation framework by injecting an adversarial network. As shown in Fig. 1, our proposed framework also involves a discriminator to further improve the segmentation performance in two folds. On the one hand, the discriminator always tries to distinguish the predicted mask and the real mask so that the whole system enforces the predicted mask to be aimilr with the real mask. On the other hand, we further take advantage of output probability (p) of discriminator to form a sample importance dispatcher, and use the generated sample importance to form a better Dice loss function as shown in the following equation:
\({L_{dice}} = \sum\limits_{i = 1}^M {{W_i}\left( {1 - 2\frac{{\sum\nolimits_{l = 1}^C {{\pi _l}\sum\nolimits_n {{r_{\ln }}{p_{\ln }}} } }}{{\sum\nolimits_{l = 1}^C {{\pi _l}\sum\nolimits_n {{r_{\ln }} + {p_{\ln }}} } }}} \right)} \)
Where \(M\) is the number of images, \(W\) is the generated sample importance, \(C\) is the number of categories, \(n\) indexes the image elements, \(r\) is the ground-truth map, \(p\) is the predicted probability map, and \(\pi \) is the assigned category weight.
Results
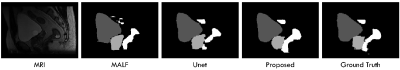
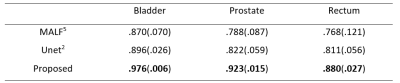
We conduct experiments on a 13-subject dataset, each containing a pair of MRI and ground-truth map. The experiments are conducted in a leave-one-out cross-validation fashion. To qualitatively demonstrate the advantage of the proposed method on this dataset, we first show the segmentation results of different tissues for a typical subject in Fig. 2. To quantitatively evaluate the segmentation performance, we use Dice ratio to measure the overlap between automatic and manual segmentation results. We report the segmentation performance in Table 1. We can observe that our proposed method significantly outperforms other approaches (p=0.00001). Specifically, our method can achieve average Dice ratios of 0.976 for bladder, 0.923 for prostate, and 0.880 for rectum. In contrast, one of the state-of-the-art segmentation methods, i.e., Unet, achieved the overall Dice ratios of 0.896, 0.822, and 0.811 for bladder, prostate and rectum, respectively, showing the superiority of our proposed approach.Conclusion
In this abstract, we have presented a novel sample attention based stochastic connection networks (SASCNet) to jointly segment pelvic organs from MRI. Specifically, the stochastic connection strategy is used to effectively address the overfitting problem of the complex network in the FCN. Moreover, the adversarial networks can further correct the segmented organs for improvement of the segmentation accuracy. More importantly, the proposed adversarial sample attention mechanism is able to improve the training. By integrating all these components into the FCN, our proposed SASCNet can achieve significant improvement in terms of both accuracy and robustness.Acknowledgements
This work was supported in part by the National Institutes of Health grants CA140413.References
1. Long, J., E. Shelhamer, and T. Darrell, Fully convolutional networks for semantic segmentation. arXiv preprint arXiv:1411.4038, 2014.
2. Ronneberger, O., P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. in International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015. Springer.
3. Srivastava, N., Hinton, G. E., Krizhevsky, A., Sutskever, I., & Salakhutdinov, R. (2014). Dropout: a simple way to prevent neural networks from overfitting. Journal of machine learning research, 15(1), 1929-1958.
4. Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., ... & Bengio, Y. (2014). Generative adversarial nets. In Advances in neural information processing systems (pp. 2672-2680).
5. Iglesias, Juan Eugenio, and Mert R. Sabuncu. Multi-atlas segmentation of biomedical images: a survey. Medical image analysis 24.1 (2015): 205-219.
6. Nie, D., Wang, L., Gao, Y., & Sken, D. (2016, April). Fully convolutional networks for multi-modality isointense infant brain image segmentation. In Biomedical Imaging (ISBI), 2016 IEEE 13th International Symposium on (pp. 1342-1345). IEEE.
Figures