3500
How easily can an existing stroke outcome deep learning model become attuned to new acquisition protocols and patient cohorts?1Center of Functionally Integrative Neuroscience and MINDLab, Department of Clinical Medicine, Aarhus University, Aarhus, Denmark, 2Cercare Medical, Aarhus, Denmark, 3Department of Neurology, Stanford University, Stanford, CA, United States, 4Department of Radiology, Stanford University, Stanford, CA, United States
Synopsis
Acute ischemic stroke is a major disease and one of the leading causes of adult death and disability. Final outcome prediction is hampered by the heterogeneity and physiological complexity of stroke progression. Convolutional neural networks have shown promising results in final outcome predictions. However, less attention has been paid to the generalizability of the results across patient cohorts. We test the applicability of an existing neural network trained on two clinical studies to completely independent cohort from the DEFUSE 2 trial. We examine how a few additional patients can be used to obtain performance comparable to the original studies.
Introduction:
Acute ischemic stroke is one of the leading causes of adult death and disability in the western part of the world. Stroke progression is heterogeneous, physiologically highly complex, and quick treatment initialization is crucial for good outcome prognosis. Accurate predictions of final tissue outcome via automatic methods will be an important tool for neuroradiologists treating stroke patients. The deep learning methods known as convolutional neural networks (CNN) has recently proved to be powerful tools in final outcome prediction. However, it is unclear how well such networks generalize to new data sets with different treatment protocols, which at the same is decisive for widespread use of such technology. In this project, we aim to investigate how a CNN perform on a new data set from facilities unknown by the original network and how a few patients can mediate the performance.Methods:
Two data sets were used: Dataoriginal: 229 patients from the I-KNOW multicenter1 (112) and perconditioning2 (117) studies. The mean of the follow-up infarct volume was 22 ml. Further details are listed in Hansen et al3. Datanew: 34 patients from the DEFUSE 2 study4. The mean of the follow-up infarct volume was 121 ml. Further details are listed in Lansberg et al4. For both data sets, patients with acute diffusion-weighted imaging (DWI), perfusion-weighted imaging (PWI) and T2-weighted-Fluid-Attenuated Inversion Recovery (T2-FLAIR) and follow-up T2-FLAIR with manual final infarct delineation were included. We compared two different models based on a CNN5 consisting of 37 layers (presented at ISMRM 20176). The first model (CNN0) was trained on 194 patients from Dataoriginal. The second model (CNN10) used CNN0 as a starting point and was then post-trained on 10 patients from Datanew for approximately 2.5 hours. The predictions were based on T2-FLAIR, trace DWI, apparent diffusion coefficient (ADC) and the following perfusion biomarkers: mean transit time (MTT), cerebral blood volume (CBV), cerebral blood flow (CBF), cerebral metabolism of oxygen7 (CMRO2), relative transit time heterogeneity (RTH), and the bolus delay estimated directly by the vascular model. The CNN0 and CNN10 models were evaluated on 24 independent patients from Datanew. The area under the receiver operation characteristic curve (AUC), the dice coefficient, and comparison between follow-up and predicted final infarct volume was evaluated and compared using Wilcoxon signed-rank tests (predictive maps are thresholded at 0.7). Moreover, the models were tested on 35 patients from Dataoriginal and the AUCs were compared through a Wilcoxon signed-rank test.Results:
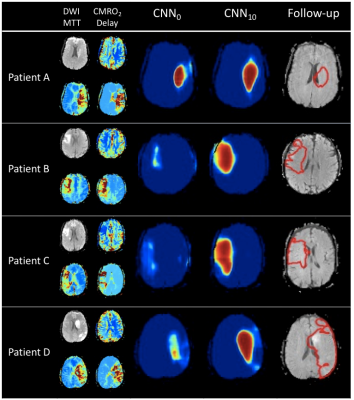
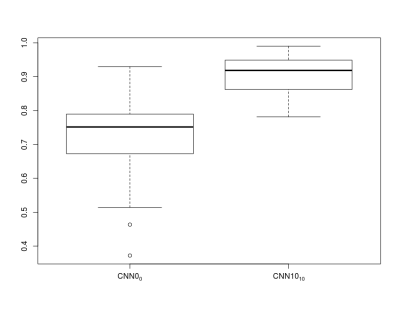
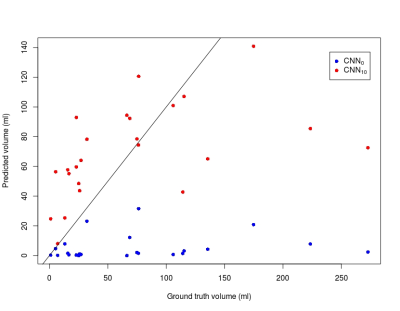
Figure 1 shows examples from the predictive models. For patient A, the predictions from the two models are similar whereas CNN0 underestimates the final infarct in the other cases. Both models underestimate patient D’s final infarct. Figure 2 shows boxplots of AUCs for the predictive models. The highest AUC was obtained for CNN10 (0.91±0.06) with a significant difference (p<0.0001) compared to CNN0 (0.71±0.13). For the dice coefficients, the difference was significant as well (p<0.0001), with CNN10 (0.35±0.19) being superior to CNN0 (0.03±0.06). As illustrated in Figure 3, the predicted volume of CNN10 is closer to the ground truth volume (mean of difference: 42.8 ml, std: 42.8) compared to CNN0 (mean of difference: 66.3 ml, std: 70.1). Both models underestimate the largest lesions. For reference, patient D from Figure 1 represents the right-most dot in Figure 3. Comparison of the models using test data from Dataoriginal showed no significant difference in AUCs (CNN0: 0.93±0.1, CNN10: 0.92±0.9, p=0.12), or in dice coefficient (CNN0: 0.39±0.28, CNN10: 0.28±0.19, p=0.08).Discussion:
We show a significant performance improvement on Datanew when the predictive model was post-trained on a few patients. We speculate this difference to be caused by the substantial difference in follow-up infarct volumes between the two data sets. The post-training approach was fast and can lead to better site-specific predictions for the CNN models’ performance which are known to be very data dependent. Our results show the importance of including a large variety of data during the training process and how a few additional cases can improve performance. A few patients from Datanew have larger final infarct lesions than the hypoperfused area on the acute scan (e.g. patient D in Figure 1). This might be due to later occlusions and as a result both predictive models underestimate these lesions due to lack of information.Conclusion:
Predictive performance of CNNs on new data sets can be improved by post-training on a few patients. This will improve final outcome prediction and generalization of CNNs yielding better automated decision support systems providing recommendations for personalized treatment and thereby hopefully better outcome for the individual patient.Acknowledgements
No acknowledgement found.References
1. I-KNOW. Integrating information from molecules to man: Knowledge discovery accelerates drug development and personalized treatment in acute stroke. 2006
2. Hougaard KD, Hjort N, Zeidler D, Sorensen L, Norgaard A, Thomsen RB, et al. Remote ischemic perconditioning in thrombolysed stroke patients: Randomized study of activating endogenous neuroprotection - design and mri measurements. Int J Stroke. 2013;8:141-146
3. Hansen MB, Nagenthiraja K, Ribe LR, Dupont KH, Ostergaard L, Mouridsen K. Automated estimation of salvageable tissue: Comparison with expert readers. J Magn Reson Imaging. 2016;43:220-228
4. Lansberg MG, Straka M, Kemp S, Mlynash M, Wechsler LR, Jovin TG, et al. Mri profile and response to endovascular reperfusion after stroke (defuse 2): A prospective cohort study. Lancet Neurol. 2012;11:860-867
5. Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. ArXiv e-prints. 2015;1511
6. Nielsen A, Hansen MB, Mouridsen K, Boldsen JK. Deep learning: Utilizing the potential in data bases to predict individual outcome in acute stroke. ISMRM. 2017
7. Jespersen SN, Østergaard L. The roles of cerebral blood flow, capillary transit time heterogeneity, and oxygen tension in brain oxygenation and metabolism. J Cereb Blood Flow Metab. 2012;32:264-277
Figures