3494
Automatic Segmentation of Carotid Vessel Wall Using Convolutional Neural Network1Electrical Engineering, University of Washington, Seattle, WA, United States, 2Radiology, University of Washington, Seattle, WA, United States, 3Radiology, Renji Hospital, Shanghai, China, 4Surgery, University of Washington, Seattle, WA, United States
Synopsis
Accurate vessel wall segmentation on black-blood MRI is an important but difficult task. Using previously annotated carotid vessel wall contours by human reviewers, a convolutional neural network (CNN) was trained to predict vessel wall region from the combination of T1-weighted and time-of-flight images. Compared with human segmentation results, the CNN-based model achieved a Dice similarity coefficient of 0.86±0.06 and a correlation coefficient of 0.96 (0.94, 0.97) in measuring vessel wall area. Fast and accurate vessel wall segmentation may help fully realize the potential of vessel wall MRI in monitoring atherosclerosis progression or regression in serial studies and clinical trials.
INTRODUCTION
Black-blood vessel wall MRI has allowed for direct visualization of atherosclerotic changes in most major arterial beds 1,2. As a technique that depicts plaque burden in 3D space and is noninvasive and free of ionizing radiation, vessel wall MRI is ideally suited for monitoring atherosclerosis progression or regression in serial studies and clinical trials. Quantitative assessment requires vessel wall segmentation. Most previous studies relied on manual segmentation 3,4, which is tedious and subject to reader variability 5. Automatic methods based on deformable models have been developed 6,7, but the intrinsic assumptions limit the performance in segments with irregular vessel wall geometries (e.g. near bifurcations or advanced plaques). Recently, neural networks that automatically learn representations of data with multiple levels of abstraction have shown advantages over traditional methods in segmentation tasks 8,9. We studied the potential utility of convolutional neural networks (CNN) in vessel wall segmentation on carotid MRI.METHODS
Patient studies
Both healthy volunteers and patients with high risk for atherosclerotic cardiovascular disease were enrolled in a clinical case-control study. Multi-contrast carotid MRI was performed on a 3T scanner (Philips Achieva) with an eight-channel carotid coil (Shanghai Chenguang) and analyzed by experienced readers. Lumen and outer wall boundaries of bilateral carotid arteries were manually traced. For this study, only T1-weighted (T1w) and time-of-flight (TOF) images were used as they are the primary weightings taken into account in manual segmentation. Imaging parameters were: 1) T1-weighted turbo SE: TR/TE = 800/10 ms, NEX =1, ETL = 10; 2) 3D TOF: TR/TE = 20/5 ms, flip angle = 20°. Spatial resolution was 0.54×0.54×2 mm3 in all images. IRB approval and informed consent were obtained.
Data pre-processing
The centerline of each carotid artery was traced by an automatic tracing method 10. Based on the centerline positions, a 64×64 window was positioned in all slices of T1w and TOF images to acquire a region of interest. Manual contours of the vessel wall were transformed into labeled images. All cases with manual segmentation results were randomly separated into: 1) training set (80%): 101 arteries (1519 slices); 2) validation set (10%; for tuning model parameters): 13 arteries (192 slices); 3) test set (10%; for evaluating model performance): 13 arteries (200 slices).
Network Model
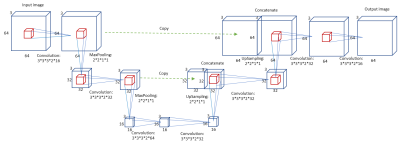
We designed a CNN model based on the U-net structure 11. Considering that human readers take into account adjacent slices during manual segmentation, we applied a modified 3D version of U-net with an input/output dimension of 64×64×3. The network structure is shown in Figure 1.
Training Process
We applied data augmentation in a combination of rotation, flipping, and shifting. Training was stopped when validation loss did not improve for 800 consecutive iterations. The model was trained on a workstation with NVIDIA GeForce GTX TITAN Xp.
Prediction and Evaluation
After centerline tracing, images were automatically cropped into the same size as the training set before vessel wall prediction. After prediction, a probability threshold, optimized in the validation set, was used to convert the probability image into a binary image. Then, the largest connected component was defined as the vessel wall region. Performance was evaluated by the Dice similarity coefficient (DSC), defined as:
$$DSC=\frac{A\cap B}{A+B}$$
where A is the labeled image and B is the segmentation result. DSC values > 0.7 indicates excellent agreement 12. The correlation coefficient between manual and automatic measurements of vessel wall area was calculated using the Pearson product-moment correlation method.
RESULTS
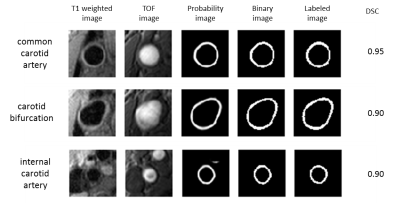
Model training took 37 minutes. A probability threshold of 0.42 was determined from the validation set. Figure 2 shows prediction results at three positions (common carotid, bifurcation, and internal carotid) in a test case. The average DSC of all 200 slices in the test set was 0.86±0.06. The correlation coefficient between manual and automatic measurements of vessel wall area was 0.96 (95% CI: 0.94, 0.97).DISCUSSION and CONCLUSIONS
In this study, fully automatic vessel wall segmentation was achieved with a high accuracy by using a CNN model that combines localization and conceptual information through reducing and restoring resolution in the U-net structure. The model appeared to be able to effectively learn the intricate patterns of the training data. Without a priori assumptions, the CNN-based segmentation method maintained excellent performance in segments of various vessel wall geometries including the common carotid artery, carotid bifurcation, and internal carotid artery segments. In the future, expanding training data to encompass a wide spectrum of atherosclerosis may further improve the performance and generalizability of the algorithm. Furthermore, it may be adapted to 3D vessel wall MRI to quantify atherosclerosis burden in tortuous arteries, where manual segmentation is difficult or unaffordable.Acknowledgements
This research is supported by grants from the National Institutes of Health (R01 HL103609) and Philips Healthcare.
We are grateful to the support of NVIDIA Corporation with donation of the Titan Xp GPU.
References
1. Dieleman N, Van Der Kolk AG, Zwanenburg JJM, et al. Imaging intracranial vessel wall pathology with magnetic resonance imaging current prospects and future directions. Circulation. 2014;130(2):192-201. doi:10.1161/CIRCULATIONAHA.113.006919.
2. Yuan C, Parker DL. Three-Dimensional Carotid Plaque MR Imaging. Neuroimaging Clin N Am. 2016;26(1):1-12. doi:10.1016/j.nic.2015.09.001.
3. Kawahara T, Nishikawa M, Kawahara C, Inazu T, Sakai K, Suzuki G. Atorvastatin, Etidronate, or Both in Patients at High Risk for Atherosclerotic Aortic Plaques: A Randomized, Controlled Trial. Circulation. 2013;127(23):2327-2335. doi:10.1161/CIRCULATIONAHA.113.001534.
4. Migrino RQ, Bowers M, Harmann L, Prost R, LaDisa JF, Jr. Carotid plaque regression following 6-month statin therapy assessed by 3T cardiovascular magnetic resonance: comparison with ultrasound intima media thickness. J Cardiovasc Magn Reson. 2011;13(1):37. doi:10.1186/1532-429X-13-37.
5. Wasserman BA, Astor BC, Sharrett AR, Swingen C, Catellier D. MRI measurements of carotid plaque in the atherosclerosis risk in communities (ARIC) study: Methods, reliability and descriptive statistics. J Magn Reson Imaging. 2010;31(2):406-415. doi:10.1002/jmri.22043.
6. Gao S, van ’t Klooster R, Brandts A, et al. Quantification of common carotid artery and descending aorta vessel wall thickness from MR vessel wall imaging using a fully automated processing pipeline. J Magn Reson Imaging. 2017;45(1):215-228. doi:10.1002/jmri.25332.
7. Yuan C, Lin E, Millard J, Hwang JN. Closed contour edge detection of blood vessel lumen and outer wall boundaries in black-blood MR images. Magn Reson Imaging. 1999;17(2):257-266. doi:10.1016/S0730-725X(98)00162-3.
8. Pinto A, Alves V, Silva CA. Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images. IEEE Trans Med Imaging. 2016;35(5):1240-1251. doi:10.1109/TMI.2016.2538465.
9. Kleesiek J, Urban G, Hubert A, et al. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. Neuroimage. 2016;129:460-469. doi:10.1016/j.neuroimage.2016.01.024.
10. Chen L, Mossa-Basha M, Balu N, et al. Development of a quantitative intracranial vascular features extraction tool on 3DMRA using semiautomated open-curve active contour vessel tracing. Magn Reson Med. 2017. doi:10.1002/mrm.26961.
11. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015:1-8. doi:10.1007/978-3-319-24574-4_28.
12. Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26(3):297-302. doi:10.2307/1932409.