3493
Predicting Contrast Agent Enhancement with Deep Convolution Networks1Radiology, Stanford University, Stanford, CA, United States
Synopsis
In this study, we tested whether deep convolutional neural networks (CNNs) could predict what an image would look like if a contrast agent was injected in the body. We trained a network to use information contained in a non-contrast MR brain exam and create a synthetic T1w image acquired after gadolinium injection. Multiple datasets including patients with tumors were used for training. Great similarities were found between the predicted and the actual images acquired after contrast agent injection. If further validated, this approach could have great clinical utility in patients who cannot receive contrast.
Introduction:
In this study, we tested whether deep convolutional neural networks (CNNs) could predict what an image would look like if a contrast agent was injected in the body. We trained a network to use information contained in a non-contrast multiparametric MR brain exam (T1w, T2w, T2*w, DWI) and create a synthetic T1w image acquired after gadolinium injection. Multiple datasets were used for training; including different numbers of patients with brain tumors and multiple combinations of pre-contrast MR parameters.Materials and Methods:
The study was approved by the local IRB committee. MR acquisitions were performed at 3T (GE Healthcare Systems, Waukesha, WI) with an 8-channel GE head coil. The MR protocol included 5 sequences (3D IR-prepped FSPGR T1w, T2w, FLAIR T2w, diffusion-weighted imaging (DWI) with 2 b values (0-1000), and T2*w) acquired before injection of 0.1 mmol/kg gadobenate dimeglumine (Multihance; Bracco) and one sequence (3D IR-prepped FSPGR T1w) acquired after injection. Data from the scanner were imported into Matlab (MathWorks Inc., Natick, MA, USA) and SPM12 was used for co-registration of the scans to the MNI template with 2mm isotropic spatial resolution.
A deep convolutional-deconvolutional neural network1 was trained to transform the multi-contrast MRI patches acquired before contrast agent injection (input) into the T1w post contrast image (output). 84 patients were scanned and 3 datasets were eventually used for training: (1) 70 patients chosen randomly in the cohort; (2) 50 patients chosen randomly in the cohort; (3) 20 patients presenting with brain tumor with obvious contrast enhancement on the T1w post image. The neural network was also trained using different combination of pre-contrast images (T1w+T2w, T1w+T2w+DWI, T1w+T2w+DWI+T2w*) to examine the type and amount of data needed to predict contrast enhancement.
Results:
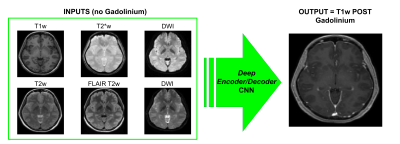
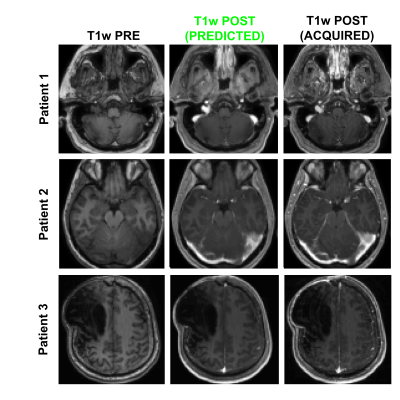
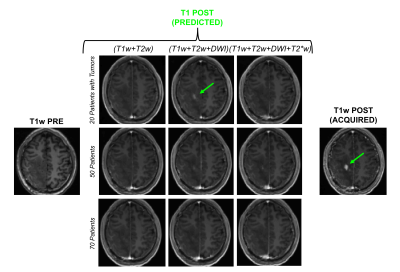
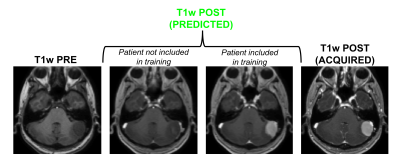
Figure 1 shows an example of training dataset (1 patient, 1 slice) that includes the 6 pre-contrast images used as input and the T1w image acquired after injection as output. In Figure 2, we show the results obtained after training (in this case, 70 patients included as well as T1w+T2w+FLAIR T2w+DWI+T2*w images), when the CNN is applied in 3 test patients not included in the network training. One can appreciate the similarities between the images predicted by the network and the actual images acquired after contrast agent injection. Although the algorithm tends to smooth the data, the global contrast in tissues is respected and all the large vessels structures (large arteries, sagittal sinus, etc.) are clearly enhanced. In patient 3, a tumor is visible in pre-contrast T1w but does not show contrast enhancement in either the predicted or acquired images. On the contrary, Figure 3 shows a case in a different patient where a portion of the tumor enhances after gadolinium injection. Here, the results obtained with different training datasets (number of patients + type of acquisitions) are also presented. The best predicted image is not obtained when the network is trained on all the patients. The best scenario is obtained when the model is trained on only the 20 patients with enhancing tumors and when T2*w data are excluded (green arrow on Figure 3). Finally, Figure 4 illustrates the difference that exists in image prediction when a patient has been seen by the network during training or not. The lack of clear enhancement in the second case suggests that better results could be obtained if more patients with similar structures had been included during training.Conclusion:
Our study suggests that CNNs are capable of synthesizing post-contrast T1w images from a combination of pre-contrast MR images. The results also indicate that better performance could be obtained if more patients (particularly with enhancing lesions) are included during training. Predicting images with higher spatial resolution and including perfusion data from Arterial Spin Labeling (ASL) or Quantitative Susceptibility Mapping (QSM) could be considered. If further validated, this approach could have a great clinical utility in patients who cannot receive contrast.Acknowledgements
Supported in part by (NIH 5R01NS066506, NIH 2RO1NS047607, NCRR 5P41RR09784).References
[1] Xie, Junyuan, Linli Xu, and Enhong Chen. "Image denoising and inpainting with deep neural networks." Advances in Neural Information Processing Systems. 2012.Figures