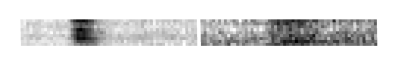3492
Perfusion MRI in stroke as a regional spatio-temporal texture1CREATIS UMR 5220, U1206, University of Lyon, Lyon, France, 2LARIS, UMR INRA IRHS, Université d'Angers, Angers, France
Synopsis
We tackle the clinical issue of predicting the final lesion in stroke from early perfusion magnetic resonance imaging. We demonstrate here the value of exploiting directly the raw perfusion data by encoding the local environment of each voxel as a spatio-temporal texture. As an illustration for this approach, the textures are characterized with Haralick coefficients computed on co-occurrence matrices and a standard support vector machine classifier is used for the classification. This simple machine learning classification scheme demonstrates good results while working on raw perfusion data.
INTRODUCTION
We address the medical issue of predicting the final ischemic stroke lesion which is still an open question 1. The ISLES challenge 2 is a testimony to the current interest of the research community for this prediction task. In this context, early perfusion magnetic resonance imaging is often used for the prediction of stroke lesion. This imaging technique produces 3D+time images which are known to be challenging 3 and the classical approaches consist in temporal or spatial preprocessing to improve the temporal signal at the pixel scale before performing prediction. In this communication, we investigate the value of raw data when encoding the regional perfusion environment as a spatio-temporal texture at the scale of patches larger than a pixel.METHODS
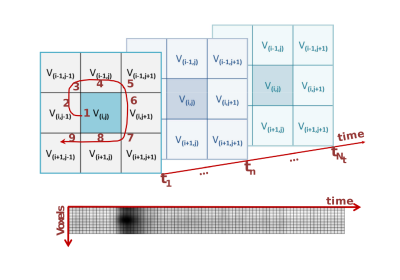
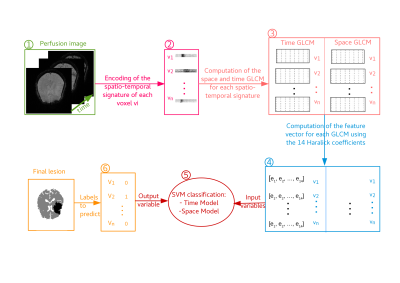
The spatio-temporal encoding of perfusion MRI sequences (Figure 1) highlights the regional signature of each voxel as a different texture when centered in regions where the blood perfusion is impaired by comparison with healthy regions (Figure 2). For illustration of this approach, these textures are characterized here with the classical Haralick coefficients 4 computed on a co-occurrence matrix coupled to a standard support vector machine 5 following the pipeline of Figure 3.RESULTS
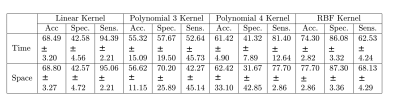
We work on longitudinal data from four patients affected by an ischemic stroke of the anterior circulation. Those four patients did not receive any thrombolytic treatment and did not reperfuse. The brain regions exhibiting a pathological hemodynamic behavior in the acute stage are therefore highly susceptible to form the final ischemic lesion. The MRI sequences used here were acquired with a 1.5 Tesla MRI scanner. The imaging protocol, approved by the ethics committee, included at admission, within three hours of stroke onset, (1) a perfusion-weighted MRI with a bolus injection of gadopentetate dimeglumine (0.1 mmol/kg), (2) a diffusion-weighted MRI and (3) a T1-weighted MRI. Also, follow-up examinations at 1 month of stroke onset were acquired, notably a FLAIR-MRI. The prediction of the fate of voxel from the perfusion MRI can thus be seen as a binary classification from the raw perfusion data encoded according to the pipeline of Figure 3. For each patient, we randomly selected the same number of non-infarcted voxels as infarcted voxels. The non-infarcted voxels were selected in the brain tissues (voxels constituted in majority of white or grey matter according to the partial volume maps) using morphomathematic approaches to determine the voxels in the neighborhood of the final lesion and those in the contra-lateral hemisphere. A total of 22100 voxels, extracted from four different patients within the European I-KNOW database 6, constituted the dataset used for our pilot study here. To account for the variability of the result presented in this section we used a K-fold cross-validation validation technique. This assesses how well the SVM classifier might generalize to an independent dataset for the prediction of the tissues final state. We divided our dataset into 100 subsets of voxels and, for each possible combination, we use 99 of the subsets for the training of the SVM classifier (99% of the data) and 1 subset for testing the quality of the classification model obtained (1% of the data). The prediction results are given in the table (Figure 4) for a vertical displacement of 1 spatial pixel and a horizontal displacement of 1 temporal pixel in the texture.DISCUSSION
The obtained performance of more than 74 % of accuracy shown in the table ( Figure 4) can be considered as already good results if one keeps in mind that the work is realized on raw perfusion data and with a very simple texture descriptor only based on the 14 Haralick coefficients. Best performances were obtained with Gaussian kernel applied on the SVM by comparison with linear or polynomial fitted kernel. It also appears from the table (Figure 4) that these performances are similar when testing a vertical or a horizontal displacement. This indicates that the local spatial redundancy carries similar amount of predictability as the temporal evolution of the perfusion signal.CONCLUSION
In this pilot study, we have proposed a new approach to encode the spatio-temporal information of voxels in raw perfusion imaging applied to stroke. We have then proposed a scheme based on co-occurrence matrix coupled to a support vector machine to realize a supervised classification of pathological and healthy voxels. This approach provides promising result with a precision of classification of more than 74% on average. This is promising indeed since a large part of the literature on stroke focuses on developing pre-processing to denoise images 7 while the classification here was realized from raw perfusion information only.Acknowledgements
This work was performed within the framework of the LABEX PRIMES (ANR-1-LABX-0063) of Université de Lyon , within the program "Investissements d'Avenir" (ANR-11-IDEX-0007) conducted by the French National Research Agency (ANR).References
1. Rekik I, Allassonnière S, Carpenter T, Wardlaw J. Medical image analysis methods in MR/CT-imaged acute-subacute ischemic stroke lesion: Segmentation, prediction and insights into dynamic evolution simulation models. a critical appraisal. NeuroImage 2012;Clinical 1:164–178.
2. Maier O, Menze BH, von der Gablentz J, Häni L. et al. ISLES 2015-A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Medical image analysis 2017;35 :250–269.
3. Willats L, Calamante F. The 39 steps: evading error and deciphering the secrets for accurate dynamic susceptibility contrast MRI. NMR in Biomedicine 2013;26:913–931.
4. Petrou M, Sevilla PG. Image processing: dealing with texture. Chichester : Wiley, 2006.
5. Bishop CM. Pattern recognition and machine learning. Springer, 2006.
6. Frindel C, Rouanet A, Giacalone M, Cho TH et al. Validity of shape as a predictive biomarker of final infarct volume in acute ischemic stroke. Stroke 2015;46(4):976-981.
7. Frindel C, Robini MC, Rousseau D. A 3-D spatio-temporal deconvolution approach for MR perfusion in the brain. Medical Image Analysis 2014;18:144-160.
Figures