3490
MR Image Synthesis Using A Deep Learning Based Data-Driven Approach1Department of Radiology, University of Wisconsin-Madison, Madison, WI, United States
Synopsis
In this study, we demonstrate MR image synthesis using deep learning networks to generate six image contrasts (T1- and T2-weighted, T1 and T2 FLAIR, STIR, and PD) from a single multiple-dynamic multiple-echo (MDME) sequence. A convolutional encoder-decoder (CED) network was used to map axial slices of the MDME acquisition to the six different image contrasts. The synthesized images provide highly similar contrast and quality in comparison to the real acquired images for a variety of brain and non-brain tissues and demonstrate the robustness and potential of the data-driven deep learning approach.
Introduction
MR image synthesis techniques allow one single image acquisition to reproduce multiple image contrasts, providing qualitative and quantitative information for research and clinical use(1). Such approaches offer the potential to dramatically reduce scan time and improve throughput. The typical synthesis procedure relies on manipulating mathematic models to convert input source MR images into a target image contrast. However, this is challenging due to the complexity of MR contrast mechanisms, image artifacts, and oversimplified model assumptions. Recently, there have been great efforts to investigate image contrast conversion between image modalities utilizing deep learning methods, with particularly successful implementations demonstrated for converting MR into CT images for PET/MR attenuation correction using convolutional neural networks(2, 3). In this study, we demonstrate the concept of MR image synthesis using deep learning networks, and emphasize this as an efficient, novel, and viable data-driven approach for image synthesis.Methods
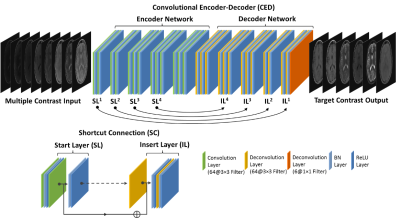
Our method utilizes the training of a deep learning convolutional neural network to store the latent spatial and contrast correlation among training images and to convert source MR images into images with target image contrast. Deep Learning Architecture: A convolutional encoder-decoder (CED) network was designed to perform the mapping function (Figure 1). More specifically, the CED network features a paired encoder network and decoder networks. The encoder consists of a set of convolution layers followed by batch normalization (BN) and ReLU activation. The decoder network is a mirrored network with the same structure as that of the encoder network but with the convolution layers substituted with deconvolution layers. Symmetric shortcut connections between layers of encoder and decoder were also added by following a full pre-activation residual network strategy to enhance mapping performance (4). Note that typical pooling and unpooling layers were removed to avoid loss of image details. Image Data: Retrospective brain MR images from 20 subjects were included in current study. Images used as target contrast include conventional 2D axial T1- and T2-weighted, T1 and T2 fluid-attenuated inversion recovery (FLAIR), short tau inversion recovery (STIR), and proton density (PD) sequences. In addition, a multiple-dynamic multiple-echo (MDME) sequence was acquired and resulted in 8 images at different TRs and TEs used as source images. Note that the MDME sequence is the same protocol as the MAGnetic resonance image Compilation (MAGiC) technique(1). All the scan was performed on a GE 3T scanner (MR750w, GE Healthcare, Waukesha, USA). Evaluation: The CED network was trained using 15 randomly selected subjects and evaluated on the remaining subjects. At the training phase, eight source images of the same slice were treated as multiple channels and combined into a single input to the network. The network was trained using mean absolute error as image loss and using an adaptive gradient-based optimization algorithm (ADAM) with an initial learning rate of 0.001 for a total of 50 epochs.Results
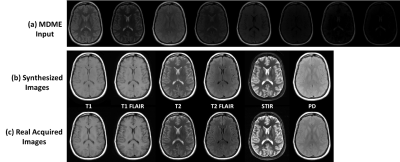
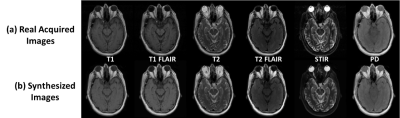
The total training phase took approximately 2 hours (computing hardware included an Intel Xeon W3520 quad-core CPU, 32 GB DDR3 RAM, and two Nvidia GeForce GTX 1080 Ti graphic cards with 7168 cores and 22GB GDDR5 RAM.). Generating synthesized MR images for one subject took less than 1 minute. Figure 2 demonstrates the synthetized MR images for T1 and T2-weighted, T1 and T2-FLAIR, STIR and PD images using our proposed method. Compared with the real acquired MR images using prescribed protocols, the synthesized images present high image quality with image features and details not visually different than ground truth. Figure 3 demonstrates another example. The synthesized images look highly similar to the real acquired for a variety of brain and non-brain tissues and demonstrate the robustness and potential of the data-driven deep learning approach.Discussion
We have demonstrated that deep learning approaches applied to MR image synthesis can produce high quality and consistent image contrast relative to MR images from real protocols. Unlike conventional synthesis methods where an explicit mathematical model is required to infer image generation, the proposed deep learning requires only sufficient data for data-driven training and thus is expected to be more robust and potentially amendable toward complex image conversion which might be challenging for simple model-based methods. Furthermore, the deep learning approach can be executed rapidly on acquired source images, making it highly suitable for clinical use. Further investigation is ongoing to fully evaluate the deep learning method with a larger dataset (including multiple disease types with both subtle and significant abnormalities) and in comparison to model-based image synthesis approaches. MR synthesis with undersampled input data is also under development to promote the image restoration function of deep learning methods.Acknowledgements
No acknowledgement found.References
1. Tanenbaum LN, Tsiouris AJ, Johnson AN, et al.: Synthetic MRI for Clinical Neuroimaging: Results of the Magnetic Resonance Image Compilation (MAGiC) Prospective, Multicenter, Multireader Trial. Am J Neuroradiol 2017; 38:1103–1110.
2. Han X: MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017; 44:1408–1419.
3. Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB: Deep Learning MR Imaging–based Attenuation Correction for PET/MR Imaging. Radiology 2017(September):170700.
4. He K, Zhang X, Ren S, Sun J: Identity Mappings in Deep Residual Networks. ArXiv e-prints 2016.
Figures