3486
Automated segmentation of abdominal organs in T1-weighted MR images using a deep learning approach: application on a large epidemiological MR study1University of Tübingen, Tübingen, Germany, 2University of Stuttgart, Stuttgart, Germany
Synopsis
In this study we implemented and validated an automated method for segmentation of T1-weighted MR images using a deep learning approach. We applied the algorithm two 80 training and 20 validation data sets drawn from an epidemiological MR study and observed high accuracy compared to manual tumor segmentation. This approach can potentially contribute to efficient analysis of large epidemiological MR studies in the future.
Introduction
Automated analysis of imaging data plays an important role with rapidly increasing numbers of performed studies and information content per study. Especially in large epidemiological imaging studies, where hundreds or thousands of individuals are scanner, visual and manual image processing is not feasible. One crucial step in the evaluation of medical image content is the recognition and segmentation of organs as a prerequisite for further analyses. The use of conventional approaches including classic image post processing or atlas-based approaches has been proposed for this task. The drawback of these approaches however is limited flexibility and generalizability. Neural networks on the other hand promise to overcome these limitations. The purpose of this study was thus to implement and evaluate deep learning - based automated segmentation of abdominal organs in MR images acquired within an epidemiological MR study.Methods
Isotropic T1-weighted, dual echo gradient echo MR datasets of 100 volunteers were obtained in the context of the KORA MR study (Cohort study in the Region of Augsburg, Germany). In all data sets, the spleen and the liver were manually segmented using the software MITK and served as ground truth for training and validation of an automated segmentation algorithm. 80 data sets were randomly selected for training and the remaining 20 data sets were chosen for validation. The segmentation algorithm used in this study is based on a combination the dense convolutional network (DenseNet) and the U-Net neural network consisting of 152 layers with a dual input scheme consisting of the fat and the water image obtained from the dual echo sequence. These dual input images were cropped to cubes of 32x32x32 voxels. The output was given as a binary mask of segmented organs. Accuracy of automated segmentation was assessed by calculating the Jaccard index between manual and automated segmentation of the respective organs. Furthermore, data sets were evaluated visually.Results
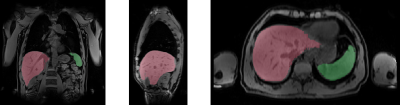
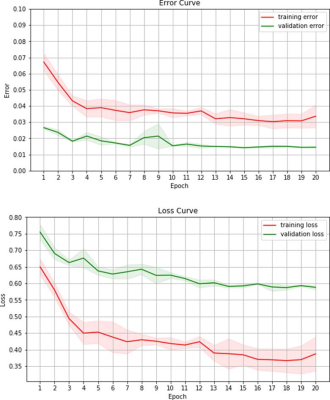
Overall, a very good agreement of automated organ segmentation was observed compared to manual segmentation. An example is shown in figure 1. On visual assessment, automated segmentation was of similar accuracy as manual segmentation for both, liver and spleen. With respect to the Jaccard index, a maximum accuracy of 98% was obtained with the optimal parameter set. Loss and error curves of training and validation passes as a function of the number of epochs are shown in figure 1.Discussion and Conclusion
In this study we investigated the possibility of automated segmentation of abdominal organs in T1 weighted MR data sets using a deep neural network. We observed high accuracy of this automated approachin comparison with manual segmentation. This methodology will be of help in the context of data analyisis in epidemiological MR studies, e.g. UK Biobank or the German National Cohort. Further organs or tissue classes can be included in the presented approach obtaining detailed automated detection and segmentation of anatomic structures. Whether the presented methodology is transferable to other data types has to be examined in further studies.
In conclusion, accurate automated segmentation of organs is feasible using the proposed deep learning approach.
Acknowledgements
No acknowledgement found.References
Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. International Conference on Medical image computing and computer-assisted intervention. 2015;9351:234–241.
G. Huang, Z. Liu, K.Q. WeinbergerDensely connected convolutional networksProceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2016), pp. 4700-4708