3485
Deep learning analysis of cardiac MRI for unsupervised classification of heart disease1Department of Computing, Imperial College London, London, United Kingdom, 2Cardiovascular Magnetic Resonance Imaging and Genetics, MRC London Institute of Medical Sciences, London, United Kingdom, 3Graduate Medical School, Duke-National University of Singapore, Singapore, Singapore
Synopsis
Magnetic resonance imaging provides detailed assessment of cardiac structure and function. However, conventional manual phenotyping reduces the rich biological information to few global metrics. A learning-based approach providing more complex phenotypic features could offer an objective data-driven means of disease classification. In this work, we exploit a convolutional variational autoencoder model to learn low-dimensional representations of cardiac remodelling which are easily visualisable on a template shape and readily applicable in classification models. This approach yielded 91,7% accuracy in the discrimination among healthy, hypertrophic and dilated cardiomyopathy subjects, and shows promise for unsupervised classification of pathologies associated with ventricular remodelling.
INTRODUCTION
Alterations in the mass or
volume of the heart define well-established classes of cardiomyopathy. However,
a learning-based approach using complex phenotypic features could offer an
objective data-driven means of disease classification [1]. While cardiovascular
magnetic resonance (CMR) allows the detailed assessment of cardiac structure
and function [2], conventional manual phenotyping reduces the rich
biological information available to a few simple volumetric parameters which
are insensitive to regional or asymmetric changes. Deep learning approaches
have recently achieved outstanding results in the medical imaging field due to
their ability to learn complex non-linear functions, but they lack
interpretability in the feature extraction and decision processes, which limits
their applicability in the clinical domain [3]. In this work, we sought
to develop a deep learning approach to capture and visualise ventricular
remodelling patterns in a dataset of images while at the same time providing high
accuracy in discriminating pathologies.
METHODS
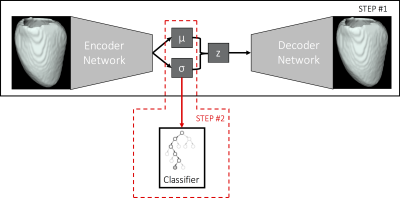
Our approach exploits a 3D
convolutional variatinal autoencoder model (CVAE) to learn a low-dimensional
representation of 3D left ventricular segmentations at end-diastole (ED) (outline
of the method in Fig. 1). The effect of each learnt latent variable can be
easily visualised on a mean template segmentation by 1) encoding the template
segmentation to the latent space, 2) varying one latent variable while keeping
the others fixed and 3) decoding the latent vector. The latent representation
learnt by the CVAE is then used as input to a random forest classifier to
discriminate between different clinical conditions.
The training set of this work
included 1,912 healthy volunteers (mean age 41±13 year, 55% females, 75%
Caucasians) from the UK Digital Heart project (UKDH). CMR was performed on a
1.5-T Philips Achieva system (Best, the Netherlands) using a high-spatial
resolution 3D balanced steady-state free precession cine sequence (60 sections,
TR 3.0 ms, TE 1.5 ms, reconstructed voxel size 1.2×1.2×2 mm). These images
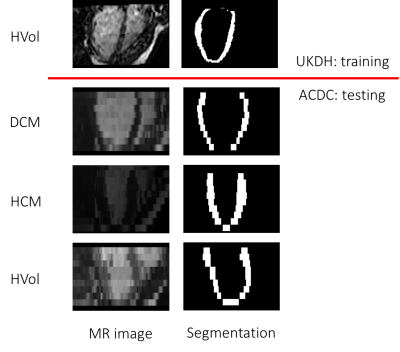
were automatically segmented and co-registered to their mean template image
[4]. The 3D left ventricular myocardium segmentations (Fig. 2) were used
to train the CVAE. As a testing set, 60 manually annotated images from the ACDC
dataset [5] (20 healthy volunteers, HVol, 20 hypertrophic
cardiomyopathy patients, HCM, and 20 dilated cardiomyopathy patients, DCM) were
employed after being rigidly registered to UKDH template image. The myocardial
ACDC shapes were presented to the trained CVAE, and their latent representation
(consisting of 64 variables) was used by a random forest classifier to classify
the three classes of subjects in a 6-fold cross-validation experiment.
RESULTS
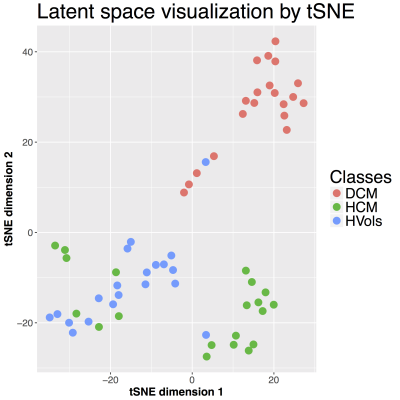
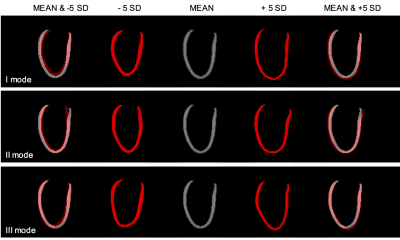
The random forest classifier achieved a mean accuracy of 91.7%. The latent 64-dimensional representation of the ACDC shapes were further reduced for visualization purposes to a bi-dimensional space using t-SNE [6]. As shown in Fig. 3, while HCM and HVols still presented some overlapping areas, DCM cases were very well clustered. In Fig. 4 the effects of the three most predictive latent features are shown on the template segmentation of the UKDH dataset. These are mainly associated with myocardium shape changes: high values of the latent variables (+5σ) corresponded to increased blood pool volume, while a smaller heart was found at -5σ.DISCUSSION
The proposed CVAE approach successfully learnt latent representations of
ventricular remodelling patterns which are readily transferable to other clinical
prediction tasks. In the clinical application shown,
the learnt representation of the ACDC dataset achieved a high classification accuracy
without exploiting any clinical variable (e.g. age or weight) and by only
analysing the ED frame. In contrast to classical end-to-end approaches, the
generative properties of the CVAE allowed the visualization of the deformation
modes associated to each latent variable. In the reported example, the network
learnt features mainly representing changes in shape and blood pool size rather
than wall thickness, as the CVAE was only trained on HVols. As a consequence,
the DCM phenotype was better clustered, as shown in the t-SNE plot (Fig. 3).
Future work will replace the random forest classifier with a neural
network and attempt to simultaneously train the classification and CVAE objectives
in order to learn more task-specific features. Further extensions will also
consider the integration of clinical variables to the latent space as well as
the inclusion of other time-points of the cardiac cycle.
CONCLUSION
We propose an unsupervised learning approach for detection of low-dimensional representations of cardiac remodelling which are informative for disease classification and easily visualisable on a template shape. In the reported application, the approach yielded high accuracy in discriminating among three clinical conditions (healthy subjects and two cardiomyopathy types). The proposed method shows promise for unsupervised classification of pathologies where ventricular remodelling has diagnostic relevance.
Acknowledgements
The authors thank our radiographers Ben Statton, Marina Quinlan and Alaine Berry; our research nurses Tamara Diamond and Laura Monje Garcia; and the study participants.References
- Konstam M.A. et al., Left Ventricular Remodeling in Heart Failure: Current Concepts in Clinical Significance and Assessment. JACC Cardiovasc Imaging. 2011 Jan;4(1):98-108
- Karamitsos, T.D. et al. The role of cardiovascular magnetic resonance imaging in heart failure. J Am Coll Cardiol. 2009 Oct;54(15):1407-24.
- Litjens, G. et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017 Dec;42:60-88.
- Bai, W. et al. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Med Image Anal. 2015 Dec;26(1):133-45.
- ACDC-MICCAI challenge dataset: http://acdc.creatis.insa-lyon.fr/
- van der Maaten, L.J.P. and Hinton, G.E. Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res. 2008 Nov;9:2579-2605.
Figures