3428
Automated breast segmentation with high reproducibility of MR-based breast density measurement1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Radiology, Stony Brook Medicine, Stony Brook, NY, United States, 3Pathology, Stony Brook Medicine, Stony Brook, NY, United States, 4Stony Brook University Cancer Center, Stony Brook, NY, United States, 5University of Arizona Cancer Center, Tucson, AZ, United States, 6Medical Imaging, University of Arizona, Tucson, AZ, United States, 7Hematology and Oncology, Stony Brook Medicine, Stony Brook, NY, United States, 8Computer Science, Stony Brook University, Stony Brook, NY, United States, 9Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States
Synopsis
Breast density(BD) is a significant risk factor for breast cancer and serves as a biomarker of risk in clinical trials. Breast segmentation is the first and an important step for accurate and reproducible BD estimation. However, the conventional manual segmentation is labor-intensive and bias-prone. Based on fat-water decomposition MRI, we developed an automated breast segmentation method and validated it against manual segmentation using 50 test-retest scans. The BD measures using our automated segmentation were very comparable to results from manual segmentation, and exhibited extremely high test-retest reproducibility. Our automated segmentation yielded more reproducible BD measures than the manual segmentation method.
Purpose
Increased breast density (BD) has been identified as a significant and independent risk factor for breast cancer1-3. Accurate measurement of BD has become a priority, not only to assess breast cancer risk but also to serve as a biomarker in longitudinal studies for evaluating the efficacy of breast cancer prevention or treatment4, 5. Fat-water decomposition MRI enables accurate BD estimation as it can truly represent the breast tissue composition in the entire breast with no ionizing radiation. Breast segmentation is the first and critical step before quantifying BD. The conventional approaches rely on manually drawn regions-of-interest (ROIs), which are cumbersome and prone to operator bias. In this study, we optimized our automated breast segmentation and validated it against manual ROIs by comparing the test-retest reproducibility of an MRI-based BD measure (MRD).Materials and Methods
50 repeated (50x2) fat-water decomposition MRI scans were performed on 31 women with early stage breast cancer enrolled in a prevention trial (24 on a 1.5T GE Signa NV-CV/I scanner using axial radial GRASE acquisition6, 26 on 3T Siemens Biograph mMR & Prisma scanners with a 3D Cartesian 6 echo GRE acquisition, both acquisition time was <5 minutes). For each repeated scan, the patient completely left the scanner and was repositioned in the scanner, re-registered and re-localized by the same technician. IDEAL fat-water separation7 was performed to generate the fat-only and water-only images, along with the fat fraction maps. Only the contralateral, unaffected breast was analyzed in this study.
Manual segmentation was performed on the water-only images by a radiology resident using MRIcron8. The ROI encompassed breast tissue and excluded skin, muscle, and nipple (approximately 30 minutes per scan). Automated segmentation was developed based on the features of fat-only and water-only images9 and with 3D regularization by applying a 3D order-statistic filter to successfully eliminate the nipple regions. An automated artifact detection was also performed to identify those slices with artifact such as fat-water swap, ghosting, etc.
MRD was calculated and calibrated to mammographic density based on our previously published technique10, which represents the actual volumetric fraction of water content in the breast after correcting the fat-water signal bias due to the intrinsic limitation of the chemical shift based fat-water separation. This MRD measure is directly comparable to mammographic density. Pearson correlation was calculated between the MRD measures from manual and automated ROIs for all 50 test scans. The test-retest reproducibility of MRD derived from manual segmentation and automated segmentation were compared using the difference between test-retest measures (∆1–2) and intra-class correlation (ICC) analyses.
Results
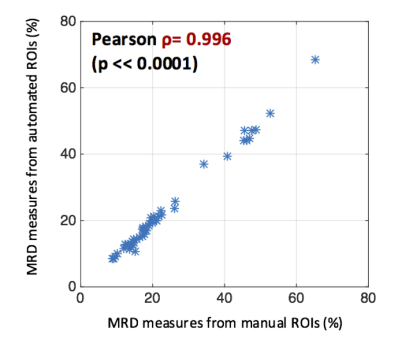
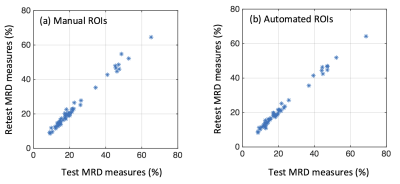
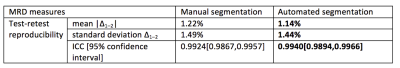
Figure 1 shows a representative fat fraction map acquired on the Biograph mMR and its corresponding manual and automated segmentation. As shown in Figure 2, BD measures of the 50 test scans using manual and automated breast ROIs were strongly correlated with Pearson ρ = 0.996, p<<0.0001. Figure 3 demonstrates the test-retest reproducibility of MRD measures generated from either manual segmentation or automated segmentation, and the statistics are illustrated in Table 1. Both manual and automated ROIs yielded highly reproducible MRD with minimal test-retest variations (1.22±1.49% vs 1.14±1.44%) and high ICCs (0.9924 vs 0.9940). However, reproducibility of the MRD obtained from automated ROIs was superior.Conclusion
Our automated breast segmentation on fat-water decomposition MRI produced very comparable BD measurements to the time-consuming and labor-intensive manual segmentation, and showed slightly higher test-retest reproducibility, which also outperformed the reported intrareader reliability of mammographic density (ICC<0.95) 11-15. This automated breast segmentation algorithm provides the basis for reproducible BD quantification for future studies and enables early detection of small changes in BD for clinical trials.Acknowledgements
This work is partially supported by NIH grants CA149417, CA161534, and Carol M. Baldwin Breast Cancer Research Fund.References
1. Boyd NF, Martin LJ, Bronskill M, Yaffe MJ, Duric N, Minkin S. Breast tissue composition and susceptibility to breast cancer. Journal of the National Cancer Institute. 2010.
2. Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MR, Giles GG, Tritchler D, Chiarelli A, Yaffe MJ. Heritability of mammographic density, a risk factor for breast cancer. New England Journal of Medicine. 2002;347(12):886-894.
3. Engmann NJ, Golmakani MK, Miglioretti DL, Sprague BL, Kerlikowske K. Population-Attributable Risk Proportion of Clinical Risk Factors for Breast Cancer. JAMA oncology. 2017.
4. Thomson CA, Chow HS, Wertheim BC, Roe DJ, Stopeck A, Maskarinec G, Altbach M, Chalasani P, Huang C, Strom MB. A randomized, placebo-controlled trial of diindolylmethane for breast cancer biomarker modulation in patients taking tamoxifen. Breast Cancer Research and Treatment. 2017:1-11. 5. Cuzick J, Warwick J, Pinney E, Duffy SW, Cawthorn S, Howell A, Forbes JF, Warren RM. Tamoxifen-induced reduction in mammographic density and breast cancer risk reduction: a nested case–control study. Journal of the National Cancer Institute. 2011.
6. Huang C, Altbach MI. Multi-Mask Multi-Seed Free Growing Field Map Estimation Algorithm for Iterative Multi-Point Water-Fat Decomposition. Paper presented at: ISMRM 17th Annual Scientific Meeting & Exhibition, 2009; Honolulu, Hawaii, USA.
7. Reeder SB, Pineda AR, Wen Z, Shimakawa A, Yu H, Brittain JH, Gold GE, Beaulieu CH, Pelc NJ. Iterative decomposition of water and fat with echo asymmetry and least‐squares estimation (IDEAL): application with fast spin‐echo imaging. Magnetic resonance in medicine. 2005;54(3):636-644.
8. Rorden C. MRICron [computer software]; 2007.
9. Rosado-Toro JA, Barr T, Galons J-P, Marron MT, Stopeck A, Thomson C, Thompson P, Carroll D, Wolf E, Altbach MI. Automated breast segmentation of fat and water MR images using dynamic programming. Academic radiology. 2015;22(2):139-148.
10. Ding J, Thompson PA, Gao Y, Marron MT, Wertheim BC, Altbach MI, Galons J-P, Roe DJ, Wang F, Maskarinec G, Thomson CA, Stopeck A, Huang C. Accurate and Reliable Fat-Water MRI Breast Density Measurements. Paper presented at: ISMRM 25th Annual Scientific Meeting & Exhibition, 2017; Honolulu.
11. Haars G, van Noord PA, van Gils CH, Grobbee DE, Peeters PH. Measurements of breast density: no ratio for a ratio. Cancer Epidemiology Biomarkers & Prevention. 2005;14(11):2634-2640.
12. Stone J, Ding J, Warren RM, Duffy SW, Hopper JL. Using mammographic density to predict breast cancer risk: dense area or percentage dense area. Breast Cancer Research. 2010;12(6):R97.
13. Sohn G, Lee JW, Park SW, Park J, Woo J, Kim HJ, Shin HJ, Kim HH, Jung KH, Sung J. Reliability of the percent density in digital mammography with a semi-automated thresholding method. Journal of breast cancer. 2014;17(2):174-179.
14. Li J, Szekely L, Eriksson L, Heddson B, Sundbom A, Czene K, Hall P, Humphreys K. High-throughput mammographic-density measurement: a tool for risk prediction of breast cancer. Breast Cancer Research. 2012;14(4):R114.
15. Eriksson L, Czene K, Rosenberg LU, Törnberg S, Humphreys K, Hall P. Mammographic density and survival in interval breast cancers. Breast Cancer Research. 2013;15(3):R48.
Figures