3427
Pipeline for Registering Histological Sections to MRI Volumes1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 3Sir Peter Mansfield Imaging Centre, School of Physics and Astronomy, University of Nottingham, Nottingham, United Kingdom, 4Insitute of Medical Informatics, Universität Lübeck, Lübeck, Germany
Synopsis
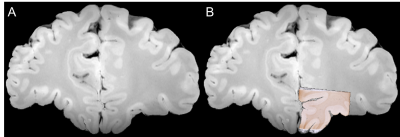
Post-mortem MRI–histology comparisons provide great potential to advance our understanding of disease, and validating the source of MRI signals, that is necessary for the development of novel imaging methods to study neurodegeneration. A semi-automated prototype of a registration pipeline is reported, that was designed for conventional sparse histological sampling. Use of the pipeline is demonstrated by inserting individual 25 x 25 mm histological sections to their respective locations in whole-brain MRI data. The registration accuracy is approximately 1 mm.
Introduction
A great deal of our current understanding of neurodegeneration lies in the post-mortem
identification of neuropathological signs, but the extent to which they can be
studied in vivo is usually limited by the sensitivity and specificity of
existing imaging biomarkers. Post-mortem MRI–histology comparisons provide
great potential to advance our understanding of disease, and validating the
source of MRI signals, that is necessary for the development of novel imaging
methods. Meaningful correlations of histological and MR data require precise
alignment of ROIs. However, it is challenged by the three-orders-of-magnitude resolution
mismatch, inherent differences in contrast generation and artefacts related to
tissue processing (tears, overlaps, debris, etc.). Furthermore, there are potentially many places in a volume of MRI where a small histological image could match,
presenting a huge search space that is likely ill-conditioned. Consequently,
most studies have either settled on manual alignment, sacrificing
reproducibility and the analysis of big data, or limit the registration problem
to isolated tissue blocks, sacrificing generality. Volumetric reconstruction
methods rely on highly demanding sequential histological sampling, which impedes
their potential wide-spread application. Here we report a semi-automated prototype
of a registration pipeline that was designed for sparse histological sampling[1], and demonstrate its use by
registering individual 25 x 25 mm histological sections to their respective
locations in whole-brain MRI data. We argue that the prototype should be
extended into a fully-automated pipeline to improve registration accuracy.Methods
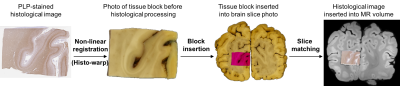
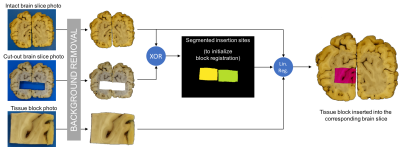
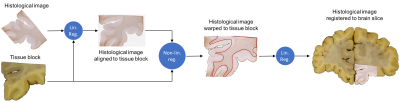
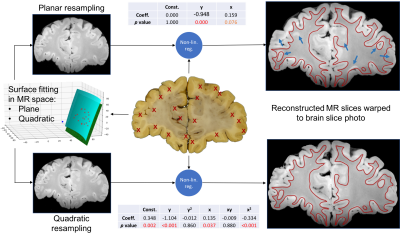
The basic registration pipeline is diagrammed in Figure 1. Subsequent figures provide detailed description of individual steps of the process (block insertion: Figure 2, histo-warp: Figure 3, slice matching: Figure 4). All operations were implemented in Python 2.7 environment, except for the non-linear registration algorithm, which runs in MATLAB.
Blue backgrounds were removed from the photos using k-means classification ($$$k=2$$$; for background and foreground) of pixels represented by RGB vectors. Through the rest of the pipeline, only the blue channel of the colour images was used, as no linear combination of colour channels in RGB and HSV space provided superior contrast between grey matter (GM) and white matter (WM).
The pipeline employs both linear (rigid-body) and non-linear (deformable) registration methods. Linear registrations are carried out by maximising the normalised mutual information[2] (NMI) of two images, using differential evolution[3]:
$$NMI=\frac{H(X)+H(Y)-H(X,Y)}{H(X)+H(Y)}$$
where $$$H\left(\cdot\right)$$$ denotes the Shannon-entropy[4] of the individual and joint histograms ($$$X$$$, $$$Y$$$) of the two images, respectively.
During non-linear registration, Gauss-Newton optimisation is used to find the deformation field $$$\mathbf{u}\left(\mathbf{x}\right):\Omega\rightarrow\mathbb{R}^2$$$ that minimises the cumulative dissimilarity index $$$\sum_{\mathbf{x}\in\Omega}{S\left(I_f,I_m,\mathbf{x}\right)}$$$ over the discrete 2-D Euclidean image domain ($$$\Omega$$$). The additional diffusion regularisation term ensures that the transformation is smooth, i.e. the displacement of pixels is confined:
$$\DeclareMathOperator*{\argmin}{arg\,min}\mathbf{\tilde{u}}\left(\mathbf{x}\right)=\argmin_\mathbf{u}{\sum_{\mathbf{x}\in\Omega}\left({S\left(I_f\left(\mathbf{x}\right),I_m\left(\mathbf{x}+\mathbf{u}\left(\mathbf{x}\right)\right)\right)^2+\frac{\alpha_1}{2}\sum_{i=1}^2{\lVert\nabla u_i\left(\mathbf{x}\right)\rVert^2}}\right)}$$
For any given deformation, the dissimilarity index of the two image functionals ($$$I_f:\Omega\rightarrow\mathbb{R}$$$, $$$I_m:\Omega\rightarrow\mathbb{R}$$$) is expressed as the sum of absolute differences between pixel-wise defined modality-independent neighbourhood descriptor (MIND)[5] vectors ($$$F_{MIND}$$$) over the search region ($$$R$$$):
$$S\left(I_f\left(\mathbf{x}\right),I_m\left(\mathbf{x}+\mathbf{u}\left(\mathbf{x}\right)\right)\right)=\frac{1}{\lvert R\rvert}\sum_{\mathbf{r}\in R}{\Big\lvert{F_{MIND}\left(I_m,\mathbf{x}+\mathbf{u}\left(\mathbf{x}\right),\mathbf{r}\right)-F_{MIND}\left(I_f,\mathbf{x},\mathbf{r}\right)}\Big\rvert}$$
$$$R$$$ is the extended neighbourhood of any $$$\mathbf{x}\in\Omega$$$, and consists of square image patches of size $$$\left(2p+1\right)^2$$$ that are centred at $$$\left\{\mathbf{r}\in R\right\}$$$. The $$$\lvert R\rvert$$$-dimensional vector $$$F_{MIND}$$$ quantifies the local self-similarity of an image by taking the sums of squared intensity differences (denoted as $$$D_p\left(\cdot\right)$$$) between corresponding pixels of the local neighbourhood ($$$\mathcal{N}$$$) and the individual patches at $$$\left\{\mathbf{r}\in R\right\}$$$:
$$F_{MIND}\left(I,\mathbf{x},\mathbf{r}\right)\propto e^{-\frac{D_p\left(I,\mathbf{x},\mathbf{x}+\mathbf{r}\right)}{\sum_{\mathbf{n}\in\mathcal{N}}{D_p\left(I,\mathbf{x},\mathbf{x}+\mathbf{n}\right)}}}\qquad\text{for}\space\mathbf{r}\in R$$
Results and Discussion
The MIND-based non-linear registration algorithm demonstrated profound robustness and excellent accuracy in the alignment of both histological (Figure 3) and MR (Figure 4) data to brain slice photos. We argue that using brain slice photos as intermediates is an essential part of the registration process that reduces the inherent ambiguity of slice-to-volume registration. Using manual input for this step and fitting polynomial surfaces to a set of corresponding anatomical locations revealed, that the geometry of brain slice photos cannot be reconstructed assuming perfectly planar cuts through the MR volume. This was supported by the significance of higher-order term coefficients of the surface fit and the visual comparison of the photo and the MR slice from planar and quadratic reconstruction (Figure 4). This irregularity of the cut surface contributes to the registration error. Treating manual entries as gold standard, the standard deviations of fitting errors (point offsets) was 1.04 mm for the planar case and 0.84 mm for the quadratic case (averages of 5 slices, i.e. 100 points), making slice-to-volume registration the bottleneck that limits the accuracy of histology-to-MR registration. Further developments are expected take place to make the pipeline fully automated and improve the registration accuracy, paving the way for a versatile tool that can be used in subsequent MRI–histology studies.Acknowledgements
Tissue was provided by the Oxford Brain Bank. Funding was provided by the MRC and Wellcome Trust. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z).References
- Pallebage-Gamarallage M, Menke RAL, Foxley S, et al. Post-Mortem Whole Brain Sampling Strategy for Histological Evaluation of Pathology Spread and Correlation with MRI Modalities in ALS.; BMC Neuroscience, 2017. Article in review.
- Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging. 2003;22(8):986-1004. doi:10.1109/TMI.2003.815867.
- Storn R, Price K. Differential Evolution – A Simple and Efficient Heuristic for global Optimization over Continuous Spaces. J Glob Optim. 1997;11(4):341-359. doi:10.1023/A:1008202821328.
- Shannon CE. A Mathematical Theory of Communication. Bell Syst Tech J. 1948;27(3):379-423. doi:10.1002/j.1538-7305.1948.tb01338.x.
- Heinrich MP, Jenkinson M, Bhushan M, et al. MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal. 2012;16:1423-1435. doi:10.1016/j.media.2012.05.008.
Figures