3424
User-defined, scanner-integrated, and real-time MRI image analysis in a cloud-based computing environment1Diagnostic and Interventional Imaging, University of Texas Health Science Center at Houston, Houston, TX, United States, 2Texas Advacnced Computing Center, University of Texas at Austin, Austin, TX, United States
Synopsis
To enhance the utility of quantitative MRI, we propose a flexible platform for high-performance cloud computing integrated with the MRI scanner. Jetstream, an NSF-sponsored open science platform for high-performance computing resources, was integrated into a clinical 3.0T MRI system for executing user-defined image analysis using the graphical pipeline environment (GRAPE) tool. Integration was achieved through the Agave platform. This framework was used for real-time quantitative T1 mapping for cartilage tissue assessment. Seamless scanner integration enabled immediate access to the results to the interpreting clinician, providing valuable quantitative information which can be incorporated in clinical practice.
INTRODUCTION
MRI can generate valuable qualitative and quantitative markers of the tissue status(1). Access to these markers can assist in identifying pathology either through visual inspection or from automated analysis. Due to the high computational load, algorithm complexity, and hardware limitations, MRI data analysis is often performed off-line, sometimes days or weeks after data acquisition. Lack of a mechanism that ties together the MRI scanner and an efficient analysis platform is thus a barrier for timely access to qualitative and quantitative MRI markers. Access to the analysis can be greatly facilitated (i) with the availability of powerful computational infrastructure, and (ii) by integrating data analysis in the scan environment.
The rise of cloud computing in recent years has provided a cost-effective solution for computationally-demanding applications. Cloud-based systems have effectively alleviated many of the problems associated with maintaining local computing infrastructure by providing scalable hardware and software as a service. MRI data analysis on cloud-based platforms has been demonstrated(2,3), but data processing is often limited to dedicated processing pipelines, and the analysis is typically performed off-line, often through a web-interface. Vendor-supplied platforms are often not cost-effective. These factors limit rapid access to the results.
We have developed a framework for bridging the MRI scanner to a cloud-based high-performance computing (HPC) cluster. This framework uses freely available software modules, and it allows the user to flexibly design novel analyses and execute them in the cloud. We further integrated this platform with a 3.0T MRI system, making the results immediately available. This framework is demonstrated for a relatively time-consuming MRI data processing task for assessing cartilage status.
METHODS
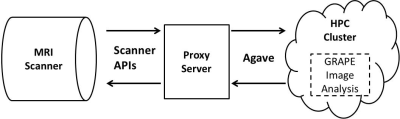
Cloud-based Real-time MRI Analysis Framework: Figure 1 illustrates the proposed framework. We used the science-as-a-service platform Agave—a representational state transfer (REST) application programming interface (API) which provides access to storage, computing, and application resources across multi-site cyberinfrastructures(4,5)—to combine a 3.0T Philips Ingenia MRI system and the Jetstream scalable cloud environment in an integrated real-time framework as recently reported(6). The interface was facilitated by a standalone workstation, the “proxy server”, used to (i) de-identify data, (ii) communicate data and instructions between the MRI scanner and the HPC cluster, and (iii) collect and organize the results. The image analysis pipelines were interactively designed using GRAPE on a desktop computer(7). GRAPE was also invoked from command line for pipeline execution on Jetstream.
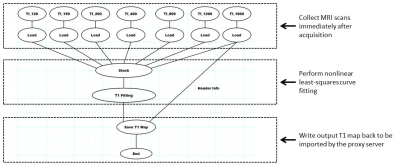
Real-time MRI Analysis Applications: Delayed gadolinium-enhanced MRI of cartilage (dGEMRIC) is a technique that depicts changes in cartilage longitudinal relaxation time (T1) associated with contrast injection as an imaging marker of cartilage degeneration(8–11). T1 determination requires a nonlinear curve-fitting that is time consuming. We developed a pipeline for same-session computation of T1 maps from 3D inversion recovery images acquired in a dGEMRIC protocol. T1 fitting used a voxel-by-voxel nonlinear least-squares fitting in GRAPE of seven echoes (inversion time (TI)=120,150,200,400,800,1200,1600ms) each with a matrix size of 384 x 384 x 13.
RESULTS
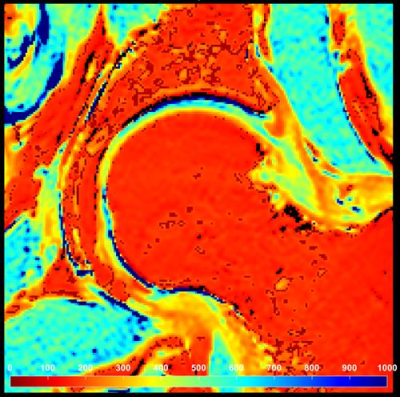
The scanner-integrated cloud-based framework for real-time MRI analysis was successful in the tested application. Images resulting from the Jetstream-GRAPE analysis were imported back to the proxy server, and subsequently to the MRI scanner. The processing time was approximately 64 seconds. A representative dGEMRIC T1 map is shown in Fig. 3.DISCUSSION
The proposed framework empowers the MRI scanner with a cloud supercomputer backend to allow the end users to have immediate access to the results and boost the use of novel and complex analysis techniques and quantitative MRI in clinical applications. The framework automates multiple tasks that would otherwise require extensive user involvement such as transferring data between the scanner and the cloud, directing data analysis, monitoring progress, error checking, and importing the results back into the scanner. With scanner integartion, the images could be effortlessly imported into the picture archiving and communication system, and would be available to the radiologist and the clinicians for assesssment. As demonstrated, the results of analysis are available on the scanner during the same imaging session, enabling the development of novel algorithms that jointly optimize data acquisition and analysis. The same “real time accessible” platform can be adapted to other complex and time-consuming computations such as nonlinear registration, segmentation, etc.CONCLUSION
Acknowledgements
This work was partially supported by the Chair in Biomedical Engineering Endowment Funds. We thank Vipulkumar Patel and Corina Donohue for assistance in conducting the MRI experiments.References
1. Tofts P. Quantitative MRI of the Brain: Measuring Changes Caused by Disease. Quantitative MRI of the Brain Measuring Changes Caused by Disease. John Wiley & Sons; 2005. 650 p.
2. Chard K, Madduri R, Jiang X, Dahi F, Vannier MW, Foster I. A cloud-based image analysis gateway for Traumatic Brain Injury research. In: Proceedings of GCE 2014: 9th Gateway Computing Environments Workshop, held in conjunction with SC 2014: The International Conference for High Performance Computing, Networking, Storage and Analysis. 2015. p. 13–6.
3. Mori S, Wu D, Ceritoglu C, Li Y, Kolasny A, Vaillant MA, et al. MRICloud: Delivering high-throughput MRI neuroinformatics as cloud-based software as a service. Comput Sci Eng. 2016;18(5):21–35.
4. Dooley R, Vaughn M, Stanzione D, Terry S, Skidmore E. Software-as-a-Service: The iPlant Foundation API. 5th IEEE Work Many-Task Comput Grids Supercomput. 2012;
5. Dooley R, Hanlon MR. Recipes 2.0: Building for today and tomorrow. Concurr Comput. 2015;27(2):258–70.
6. Allen WJ, Gabr RE, Tefera GB, Pednekar AS, Vaughn MW, Narayana PA. Platform for Automated Real-Time High Performance Analytics on Medical Image Data. J Biomed Heal Informatics. 2017; DOI: 10.1109/JBHI.2017.2771299
7. Gabr RE, Tefera GB, Allen WJ, Pednekar A, Narayana PA. GRAPE: a graphical pipeline environment for image analysis in adaptive magnetic resonance imaging. Int J Comput Assist Radiol Surg. 2017;12(3):449–457.
8. Bashir A, Gray ML, Burstein D. Gd-DTPA2- as a measure of cartilage degradation. Magn Reson Med. 1996;36(5):665–73.
9. Bashir a, Gray ML, Boutin RD, Burstein D. Glycosaminoglycan in articular cartilage: in vivo assessment with delayed Gd(DTPA)(2-)-enhanced MR imaging. Vol. 205, Radiology. 1997. p. 551–8.
10. Bashir A, Gray ML, Hartke J, Burstein D. Nondestructive imaging of human cartilage glycosaminoglycan concentration by MRI. Magn Reson Med. 1999;41(5):857–65.
11. Zilkens C, Tiderius CJ, Krauspe R, Bittersohl B. Current knowledge and importance of dGEMRIC techniques in diagnosis of hip joint diseases. Vol. 44, Skeletal Radiology. 2015. p. 1073–83.
12. Gabr RE, Pednekar AS, Govindarajan KA, Sun X, Riascos RF, Ramirez MG, et al. Patient-specific 3D FLAIR for enhanced visualization of brain white matter lesions in multiple sclerosis. J Magn Reson Imaging. Wiley Online Library; 2017;46(2):557–564.
Figures