3421
SeedNet: a sliding-window convolutional neural network for radioactive seed detection and localization in MRI1Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 2Radiation Oncology, MD Anderson Cancer Center, Houston, TX, United States
Synopsis
Radioactive seed localization is an essential step in quantifying the dose delivered to the prostate and surrounding anatomy after low-dose-rate prostate cancer brachytherapy. Currently, dosimetrists spend hours manually localizing the radioactive seeds in postoperative images. In this work, we investigated a novel sliding-window convolutional neural network approach for automatically identifying and localizing the seeds in MR images. The method doesn’t rely on prior knowledge of the number of seeds implanted, strand placements, or needle-loading configurations. In initial testing, the proposed approach achieved a recall of 100%, precision of 97%, and processing time of ~0.5-1.5 minutes per patient.
Introduction
Accurate localization of the implanted radioactive seeds relative to critical normal structures is essential in low-dose-rate (LDR) prostate cancer brachytherapy. Compared to CT, MRI provides excellent depiction of the anatomy but is limited in conclusively identifying the radioactive seeds, especially in the presence of other similar image features, such as needle tracks. Visualizing both anatomy and radioactive seeds with MRI is possible with positive-MR-contrast seed marker technology1, whereby an MRI-signal-producing solution is encapsulated and placed as the spacers between adjacent radioactive seeds. With this technology, radioactive seeds are manually identified by a medical dosimetrist. However, this process is time-consuming and is subject to human errors. In this work, we developed SeedNet: a sliding-window convolutional neural network (SWCNN) approach for automatically identifying and localizing the radioactive seeds and markers.Methods
Twenty-one post-implant patients were scanned with a 3D balanced steady-state free precession pulse sequence (CISS) on a 1.5T Siemens Aera scanner using a 2-channel rigid endorectal coil in combination with two 18-channel external array coils. The scan parameters were: TR/TE=5.29/2.31 ms, RBW=560 Hz/pixel, FOV=15 cm, voxel dimensions=0.59×0.59×1.2 mm, FA=52°, and total scan time of 4-5 minutes.
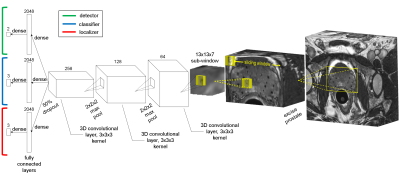
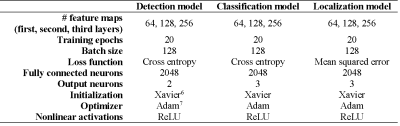
To limit the image context and computation time, a rectangular cuboid region of interest (ROI) encompassing the prostate and the implanted seeds was cropped from the images. A sliding-window algorithm was written to scan the ROI in 13x13x7-voxel sub-windows with a 2x2x1 stride. Three 3D convolutional neural networks (CNNs) were trained to perform seed and seed marker detection, classification, and localization tasks. SeedNet’s architecture (partially inspired by LeNet52, AlexNet3, and OverFeat4) is shown in Fig. 1. The main hyperparameters for constructing the networks are shown in Table 1. 2x2x2 max pooling was performed after the first two convolutional layers. Dropout5 (50%) was used after the third convolutional layer to prevent overfitting. During testing, the seed/marker inferences from the detection CNN were passed to the classification CNN, and the seed/marker inferences from the classification CNN were passed to the localization CNN. The inferences from the localization CNN were mapped back to the original image stack.
SeedNet was written in TensorFlow and trained, cross validated, and tested on a Linux RedHat v7.2 server with four NVIDIA Quadro K2200 GPUs. The CNNs were trained on sub-windows from 18 patients, 20% of which were reserved for cross validation (CV), and tested on three patients. The labeled locations of the seeds and markers of the 21 datasets were created from dosimetry plans that were previously completed by a certified medical dosimetrist. Multiple inferences for a given seed or seed marker were possible because the sliding window visited all locations within the image and therefore visited each seed/marker multiple times. The seed locations inferred from SeedNet were compared with the dosimetrist’s manually identified seed locations to determine the network’s performance.
Results
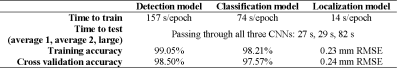
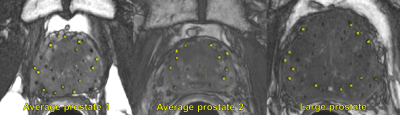
Results for training and cross validating the three networks are summarized in Table 2. The training and CV accuracies for the CNNs were: detection-99.05% (training), 98.50% (CV); classification-98.21% (training), 97.57% (CV); and localization-0.23 mm RMSE (training), 0.24 mm RMSE (CV). The total testing time was 0.5-1.5 minutes, depending on the prostate size. A few false positives (FPs) remained in dark crevices of anatomical boundaries and can be corrected with postprocessing by incorporating known information about the geometry of seed strands. Representative slices from each test patient are shown in Fig. 2. The chain of CNNs correctly identified and localized 52/52 seeds (3 FPs) in the first average-sized prostate, 56/56 seeds (3 FPs) in the second average-sized prostate, and 124/124 seeds (2 FPs) in the large prostate.Discussion
To our knowledge, SeedNet is the first application of CNNs to radioactive seed identification and localization in post-implant MR images. The high identification and localization accuracy (100% recall, 97% precision) demonstrate the potential for automated radioactive seed localization in post-implant MR images, a critical step for MR-only post-implant dosimetry. The short time (~0.5-1.5 minutes) to run the algorithm is a substantial improvement over the current manual seed identification process.Conclusion
We have presented an automated approach to radioactive seed detection and localization in MR images of the prostate after LDR brachytherapy. The algorithm does not require prior knowledge of the number of seeds implanted, strand placements, or needle-loading configurations. The algorithm is accurate and efficient, providing a substantial savings in time and cost over the current standard of manual dosimetry. The SWCNN approach presented could potentially be used in other detection/classification/localization applications in which prior information about the number/type/location of the objects in the image is unknown (e.g. a generalized detection algorithm).Acknowledgements
No acknowledgement found.References
[1] Frank SJ, Stafford RJ, Bankson JA, et al. “A novel MRI marker for prostate brachytherapy,” Int J Radiat Oncol Biol Phys 2008;71(1):5-8.
[2] LeCun Y, Bottou L, Bengio Y, Haffner P. “Gradient-based learning applied to document recognition,” Proc. IEEE 1998;86(11):2278-2324.
[3] Krizhevsky A, Sutskever I, Hinton GE. “ImageNet classification with deep convolutional neural networks,” Neural Information Processing Systems 2012.
[4] Sermanet P, Eigen D, Zhang X, et al. “OverFeat: Integrated recognition, localization and detection using convolutional networks,” arXiv:1312.6229, 2013.
[5] Hinton GE, Srivastava N, Krizhevsky A, et al. “Improving neural networks by preventing co-adaptation of feature detectors,” arXiv:1207.0580, 2012.
[6] Glorot X, Bengio Y. “Understanding the difficulty of training deep feedforward neural networks,” Proc. 13th International Conference on Artificial Intelligence and Statistics 2010, pp. 249-256.
[7] Kingma DP, Ba J. “Adam: A method for stochastic optimization,” arXiv:1412.6980, 2014.
Figures