3420
Quantitative Micro-Vasculature Volume Assessment of Intra Tumoral Susceptibility Signal (ITSS) in differentiating Grade-III from IV glioma1Centre for Biomedical Engineering, Indian Institute of Technology, Delhi, New Delhi, India, 2Philips Health Systems, Philips India Limited, Gurgaon, India, 3Department of Radiology and Imaging, Fortis Memorial Research Institute, Gurgaon, India, 4SRL Diagnostics, Fortis Memorial Research Institute, Gurgaon, India, 5Biomedical Engineering, AIIMS, New Delhi, Delhi, India
Synopsis
Angiogenesis transforms gliomas from low-to-high-grade. Vasculature-properties are of essential prognostic-value within grade-III and IV glioma as compared to grade-II. High-resolution susceptibility-weighted imaging (SWI) improves the diagnostic accuracy1. Existing Semi-quantitative methods are user-dependent which manually counts intra-tumoral-susceptibility-signal-intensities (ITSS); a combination of haemorrhage and vasculature. Haemorrhage contributes to false ITSS-count and subsequently to misclassification of tumor-grading. We propose a non-invasive segmentation-based-quantitative approach that calculates the R2-Star relaxivity maps of ITSS, automatically removes haemorrhages from ITSS based on high-R2-Star relaxivity of haemorrhage and finally calculate microvasculature volume within glioma. The proposed-method scores over the existing semi-quantitative method in-terms-of ITSS-estimation and grading-accuracy.
Purpose
Tumor growth is dependent on the dynamic process of angiogenesis which transforms gliomas from low to high-grade. Studies have shown1 that mean vessel density and vasoactive-endothelial-growth-factor (VEGF) are of essential prognostic value as they increase with increasing tumor grade. Grade-II tumor have relatively better prognosis. Within grade-III and IV high variations of biological-behavior and genetic-nature can be seen, but differentiation based on imaging techniques is relatively poor. The correct prognosis becomes necessary within grade-III and IV due to the survival rate and time constraints. Multi-parametric-imaging approach has been attempted to differentiate these two groups where high resolution susceptibility-weighted imaging (SWI) improves the diagnostic accuracy1. Park.et.al2-3 has established semi-quantitative method of manually counting shape-based intra-tumoral-susceptibility-signal intensities (ITSS) from SWI which is a combination of haemorrhage and vasculature. This count has been shown to determine the degree of ITSS as well as the glioma-grading. However, the above method is manual-count-based and highly user-dependent. Haemorrhage contains deoxyhemoglobin and hemosiderin depending upon its age whose susceptibility effects cause high signal decay resulting in high-relaxivity (R2-Star). We propose a non-invasive segmentation-based-quantitative approach that calculates the microvasculature volume within tumors by automatically filtering out the haemorrhage from ITSS based on R2-Star quantitative variations. This work evaluates the efficiency of microvasculature-volume to differentiate grade-III from grade-IV glioma viz-a-viz described method in the literature.Methods
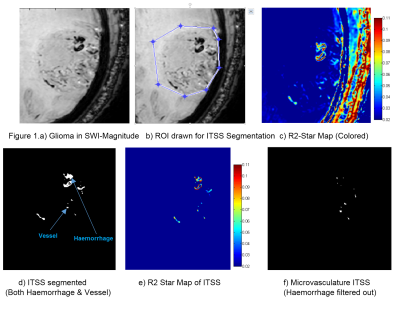
This IRB-approved retrospective study included total 20 histology-confirmed glioma patients (10 each of grade III and IV). All patients underwent conventional-MRI and SWI on a 3.0T MRI scanner (Ingenia, Philips Healthcare, The Netherlands). SWI was acquired with 4 echoes at at 5.6,11.8,18 and 24.2 ms (TR = 31ms; flip-angle 17°, slice thickness 1-mm, matrix 384×384, FOV 240×240 mm2). The SW-Magnitude images are obtained from scanner by multiplying fast-field-echo(FFE)-M image with a phase mask derived from PADRE (Phase-Difference-Enhanced-imaging) filtering process. PADRE algorithm calculates phase-mask from the homodyne-filtered phase image. The R2-Star maps were calculated using SWI-multi-echo magnitude images using below equation described by Haacke et.al4
$$R_2^* = - \frac{\sum_{m=2}^N {log(\frac{p_{m}}{p_{1}})*(TE_{m}-TE_{1})}}{\sum_{m=2}^N{(TE_{m}-TE_{1})^{2}}}$$
Thresholding and shape-based structuring element dependent algorithm developed in-house using MATLAB was used to segment ITSS from SWI-magnitude images for all the tumor slices. R2-Star values were picked for haemorrhage tissues from proven brain-haemorrhage data-pool (identified by expert radiologists) and those values were used as thresholds for detecting haemorrhage ITSS. Haemorrhages appear as large conglomerating blobs. Using this R2-Star threshold of haemorrhage a connected-component-shape-analysis segmentation algorithm was developed in MATLAB, which detects conglomerated haemorrhage ITSS blobs from all the tumor slices and filter them out. Remaining ITSS was considered to contribute to be microvasculature within the tumor. The microvasculature volume was computed over all the tumor slices through an in-house-developed algorithm. This volume calculation was done for all the 20 cases. Statistical unpaired-t-test was performed to check whether there is significant difference of microvasculature ITSS volume between grade-III and grade-IV cases. Also in-parallel ITSS-count and degree were taken based on Park-et-al’s semi-quantitative method. All these computations were finally compared with the histopathological grading results to compute accuracy of tumor-grading.
Results
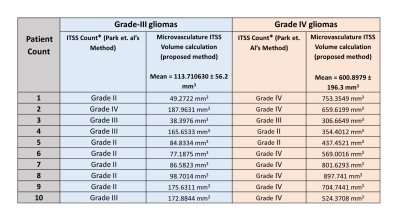
Table-1 summarizes the microvasculature ITSS volume as well as ITSS-count from grade-III and grade-IV glioma. Microvasculature volume was much higher in grade-IV compared to grade-III glioma. Unpaired-t-test between microvasculature ITSS volume of grade-III and IV provided p<0.0001. The semi-quantitative method was accurate only in detecting 30% cases of grade-III and 70% cases of grade-IV; while current method differentiated all the 20 cases correctly as grade-III or IV disease. Figure 1.Discussion and Conclusion
Aggressiveness of a tumor can truly be predicted by tumor vasculature quantification. The proposed method scores over the existing semi-quantitative method in terms of ITSS estimation and grading accuracy. It is probably due to removal of haemorrhages in current approach as haemorrhages usually contribute to false ITSS-count and subsequently to misclassification of tumor-grading. The current approachThe intra-tumoral-microvasculature is induced by the tumor-hypoxia which causes formation of the neo-vasculature and increased intra-tumoral-oxygen demand; resulting in increased deoxy-hemoglobin in the tumor-vasculature and hence the enhanced BOLD effect. This proposed quantitative approach needs further evaluation in a large sample size of these different grades of glioma to further establish the cut-off-ranges for each grade. It also has the scope of extending to a stand-alone option for quantitative and non-invasive tumor grading where ITSS is visible.Acknowledgements
The Authors acknowledge technical support of Philips India Limited and Fortis Memorial Research institute Gurugram in MRI data acquisition. Authors thank Dr. Indrajit Saha, Prof. RKS Rathore, Dr. Prativa Sahoo for technical support in data-processing. This work was supported by grant from Science and Engineering Research Board (IN) (YSS/2014/000092).References
[1] Saini J, Gupta PK, Sahoo P, Singh A, Patir R, Ahlawat S,Beniwal M, Thennarasu K, Santosh V, Gupta RK.Differentiation of grade II/III and grade IV glioma by combining “T1 contrast-enhanced brain perfusion imaging” and susceptibility-weighted quantitative imaging. Neuroradiology, 2017 (online) [https://doi.org/10.1007/s00234-017-1942-8]
[2] Pinker K, Noebauer-Huhmann IM, Stavrou I, et al. High-resolution contrast-enhanced, susceptibility-weighted MR imaging at 3T in patients with brain tumors: correlation with positron-emission tomography and histopathologic findings AJNR Am J Neuroradiol 2007;28:1280–86
[3] Park MJ, Kim HS, Jahng GH, et al. Semiquantitative assessment of intratumoral susceptibility signals using non-contrast-enhanced high-field high-resolution susceptibility-weighted imaging in patients with gliomas: comparison with MR perfusion imaging. AJNR Am J Neuroradiol Aug 2009: 30:1402– 08
[4] Haacke EM, Cheng NY, House MJ, et al. Imaging iron stores in the brain using magnetic resonance imaging. Magn Reson Imaging 2005; 23: 1–25.