3417
Collaborative volumetric magnetic resonance image rendering on consumer-grade devices1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Interactive Commons, Case Western Reserve University, Cleveland, OH, United States, 3Department of Radiology, School of Medicine, Case Western Reserve University, Cleveland, OH, United States
Synopsis
We present a system for intra- or post-acquisition 3D rendering of volumetric MRI datasets for independent or collaborative use on AR/VR and mobile platforms. Consumer-grade head mounted displays, phones, and computers are used to provide 3D visualizations. Datasets can be windowed and leveled in the same manner as classic visualizations, and arbitrary slices can be selected and viewed in real time in the context of the whole volume. Real world dimensionality and spatialization is retained. Using this system, multiple users can interact with a dataset collaboratively using current AR/VR platforms or any modern cellphone, tablet, or laptop.
Purpose
Medical imaging traditionally relies on planar representations of patient data without three-dimensional context. This is directly contrary to the reality of the human body, where spatial information and feature dimensionality are of critical importance to proper diagnosis and treatment. With the increasing availability of high quality stereoscopic head mounted displays (HMDs) as well as high performance non-stereoscopic mobile platforms, there exists a clear opportunity to “re-spatialize” medical images to enable direct and intuitive interaction both at the scanner and in the reading room.
Clinicians are accustomed to collaboratively diagnosing patients through group inspection and discussion. This process is impeded by traditional reading methods – collaboratively evaluating scans by scrolling through a DICOM stack can’t compare to physically searching for and pointing out anomalies. Moreover, a low latency real-time multi-user 3D environment presented intra-acquisition using AR, VR, or touchscreen offers a means of clinician engagement that better approximates traditional diagnostic practice. We have therefore developed a visualization pipeline to display both realtime and static MRI data on a group of HMDs or mobile devices for collaborative viewing.
Methods
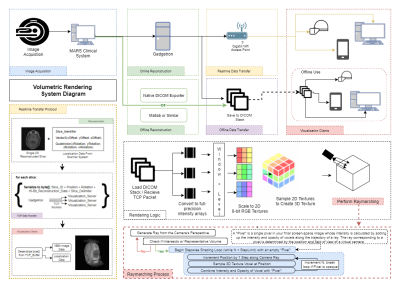
Figure 1 details the rendering pipeline and approach. Rendering clients could import data from offline-reconstructed DICOMs or receive realtime-reconstructed data over TCP. Images were loaded into a volumetric texture system without compression, allowing the user to manipulate image window and level at runtime. Voxel sizes were calculated from scanner data to allow use of any isotropic or anisotropic dataset. Rendering was accomplished by a progressive ray tracing (“raymarching”) approach (1,2) described in Figure 1. Each voxel’s intensity value contributed to the final screenspace pixel color via the raymarching graphics shader. Noise was reduced using an adjustable noise floor filter (Figure 2).
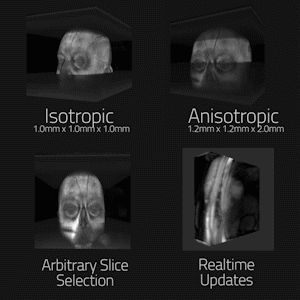
In order to test the rendering pipeline’s accuracy, two offline magnetic resonance datasets of the brain were acquired with 3D MP-RAGE sequence with isotropic spatial resolution of 1mmx1mmx1mm, and anisotropic resolution of 1.2mmx1.2mmx2mm. The datasets were reconstructed offline to DICOMs and loaded onto client systems. Initialization and framerate performance was evaluated on a Desktop PC (3.2Ghz CPU, Quadro K4200 GPU), Oculus Gear VR (Samsung Galaxy S8+), and Microsoft Hololens for multiple matrix sizes and raymarching step counts. These devices represent, from highest to lowest, the three major performance tiers for target devices. Lag in realtime transmission was tested over Gigabit Ethernet and 802.11ac Wifi using single, 3-slice, and full volume updating of a volumetric heart acquisition. (FLASH sequence w/ undersampled radial trajectory, reconstructed using through-time radial GRAPPA on Gadgetron (3,4)).
Users were provided with runtime controls for window, level, position, and orientation as well as slice plane position and orientation through handheld controllers, gestures, or touchscreen interactions. The clipping position could also be controlled by the user’s head to allow natural data exploration. In multi-user use, render parameters were synchronized so users shared the same visualization. Additional client-side rendering parameters included raymarching step count and framerate-dependent downsampling to maintain performance.
Collaborative sessions were accomplished across all tested platforms. A host device began a “reading session” which other client devices could join. Hosts retained control of rendering parameters, while all users could explore the rendering. Participants on 2D platforms could interact via touchscreen, with a projection of their simulated “position” visualized in the AR/VR “reading room” to enhance embodiment.
Results
Figure 2 is an animated GIF showing results from the rending pipeline. It also demonstrates the noise rejection filter system in use, enhancing isolation of useful data to improve the visualization. Figure 3 maps system initialization and render times versus matrix size and volume rendering stepcount, as well as transmission lag in realtime use. Though dwarfed by the Desktop PC, both mobile devices rendered standard 128^3 datasets near full fidelity with minimal loading times. Single, three-slice, and full-volume realtime updating all supported >10fps transfer. Figure 4 is an animated GIF demonstrating accurate rendering of isotropic and anisotropic MP-RAGE datasets, dynamic clipping planes, and realtime intra-acquisition updating of a rendered volume. Figure 5 is an animated gif of a collaborative session utilizing the platform.Discussion
Mixed Reality and mobile platforms offer highly flexible and collaborative methods for displaying MR datasets in full context. All three consumer-grade systems were capable of rendering standard matrix size isotropic and anisotropic acquisitions at acceptable framerates for both static and realtime acquisitions. Collaborative reading sessions of volumetric data sets allowed multiple users to simultaneously interact with a dataset from independent devices. This success indicates a need for further exploration of the impact a platform-agnostic volumetric rendering system could have on creating the three-dimensional collaborative reading rooms and interventional suites of the future.Acknowledgements
Siemens Healthcare, R01EB018108, NSF 1563805, R01DK098503, and R01HL094557.References
1. Levoy, M. Efficient ray tracing of volume data. ACM Trans. Graph. 1990; 9, 3, p.245–261
2. Zhou et al. Real-time Smoke Rendering Using Compensated Raymarching. Microsoft Research. 2007.
3. Hansen MS, Sørensen TS. Gadgetron: An open source framework for medical image reconstruction. Magn. Reson. Med. 2013;69:1768–1776.
4. Franson D, Ahad J, Hamilton J, Lo W, Jiang Y, Chen Y, Seiberlich N. Real-time 3D cardiac MRI using through-time radial GRAPPA and GPU-enabled reconstruction pipelines in the Gadgetron framework. In: Proc. Intl. Soc. Mag. Reson. Med. 25.; 2017. p. 448.
Figures