3413
Can brain MRI skull-stripping methods be further improved using manual segmentation as ground-truth for validation?1Seaman Family Magnetic Resonance Research Centre, Calgary, AB, Canada, 2Medical Image Computing Lab, Campinas, Brazil
Synopsis
Automatic skull-stripping (SS) methods have reached a high level of accuracy compared to expert manual segmentation (typically defined as the “ground-truth”), but SS is still an active research area with many methods being proposed every year. In this work, we use twelve T1-weighted brain magnetic resonance (MR) images with each image having two different manual segmentations performed by experts, and four state-of-the-art SS methods to assess if it is possible to evaluate further accuracy improvements to SS. Our results indicate that at the current level of SS accuracy, this is not possible using single expert manual segmentation.
Introduction
Skull-stripping (SS) is the process of segmenting brain from non-brain tissue. In MR images, SS is an initial step for many quantitative image analysis applications1. It is an active research field2-4 and SS performance is usually assessed using manual expert segmentation. Our goal is to show that at the current level of SS accuracy, it is not possible to assess further SS accuracy improvements using single expert manual segmentation. In other words, performance metrics are driven by both the method under evaluation as well as the comparison (“ground truth”) reference data.Materials and Methods
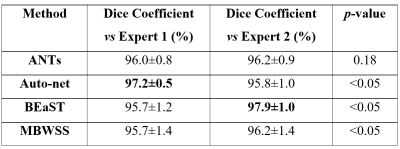
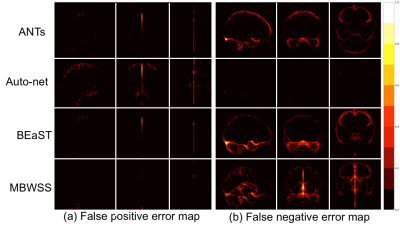
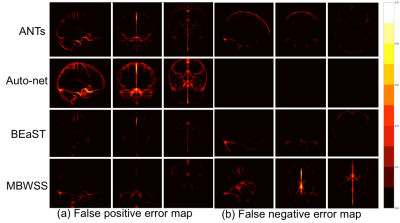
Twelve T1-weighted brain magnetic resonance (MR) volumes acquired on different scanner vendors (Siemens, Philips, General Electric) and at different magnetic field strengths (1.5 T and 3 T) were manually and independently segmented by two different experts. Two image volumes for each vendor-magnetic field strength combination were segmented. The volumes were skull-stripped using four automatic methods: 1) Advanced Normalization Tools5 (ANTs), 2) Auto-context Convolutional Neural Network (Auto-net) inspired by Salehi et al.6, 3) Brain Extraction Based on Nonlocal Segmentation Technique7 (BEaST), and 4) Marker Based Watershed Scalper8 (MBWSS). The automated results were compared against the manual segmentations using the Dice coefficient9, which is a standard performance metric for assessing agreement and ranges from 0% to 100%. Mean ± standard deviation (over subjects) was reported. A paired Student’s t-test was used to assess statistical significance with p<0.05 used to determine statistical significance. Sagittal, coronal and axial disagreement (error) maps were built by using non-linear registration and averaging the false positives and false negatives, respectively, using manual segmentation from one expert as the reference. The image volumes used in this study are publicly available as part of the Calgary-Campinas-359 dataset available at: http://miclab.fee.unicamp.br/tools.Results
The Dice coefficient results of the automatic methods are summarized in Table 1. The Dice coefficient comparing the two expert manual segmentations was of 96.4%±0.7%. The smallest and largest volume differences between the expert segmentations was 0.89% and 7.30%, respectively. Figures 1-3 illustrate the disagreement maps of the automatic methods against expert 1 manual segmentation, automatic methods against expert 2 manual segmentation, and expert 1 versus 2 manual segmentation, respectively.
Discussion
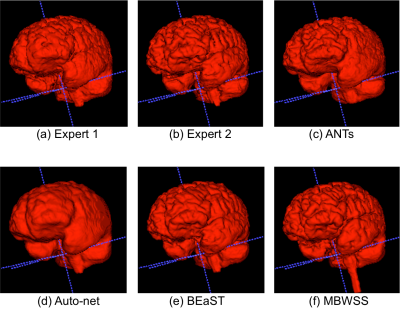
The results show that the assessment of SS methods is highly dependent of the manual segmentation used as the ground truth or reference, some methods obtained better results when compared to expert 1, while others fared better against expert 2. The ranking of the SS methods on the Dice coefficient varied with the expert segmentation reference segmentation used to compute the metrics. All methods, except ANTs, were statistically different (p < 0.05) with selected expert manual segmentation used as the reference to compute the Dice coefficient. As modern SS methods approach the accuracy of manual expert segmentation agreement (>95% for SS according to our results), it becomes challenging to objectively assess further improvements in segmentation accuracy. A possible approach to overcome this problem is to make the consensus of multiple expert manual segmentations using algorithms, such as the Simultaneous Truth and Performance Level Estimation10 (STAPLE) to try to improve the quality of the ground truth reference segmentation. Brain manual segmentation is time consuming, taking about five hours to segment a single volume11, making this approach expensive. A number of brain volumes with multiple expert segmentations would be necessary to validate automatic methods. At the current stage of SS methods, secondary criteria, such as processing time, robustness, and visual qualitative assessment (Figure 4) should also be assessed when deciding which is the most suitable SS method for a given application.Conclusions
SS methods are currently approaching the same level of accuracy as expert manual segmentation, therefore making it extremely difficult to assess algorithmic improvements in the field using conventional approaches. In light of this fact, secondary criteria are becoming more important when choosing the most appropriate SS method for a given application. We expect that this difficulty in assessing segmentation improvements extends to other segmentation problems.Acknowledgements
This project was supported by FAPESP CEPID-BRAINN (2013/075593) and CAPES PVE (88881.062158/2014-01). Roberto A. Lotufo thanks CNPq (311228/2014-3), Roberto Souza thanks FAPESP (2013/23514-0) and the NSERC CREATE I3T foundation, Oeslle Lucena thanks FAPESP (2016/18332-8). Richard Frayne is supported by the Canadian Institutes of Health Research (CIHR, MOP-333931) and the Hopewell Professorship in Brain Imaging.References
1. Smith SM. Fast robust automated brain extraction. Human brain mapping 2002;17(3):143-55
2. Benson C, Lajish V, Rajamani K. A Novel Skull Stripping and Enhancement Algorithm for the Improved Brain Tumor Segmentation using Mathematical Morphology. International Journal of Image, Graphics and Signal Processing 2016;8(7):59
3. Kleesiek J, Urban G, Hubert A, et al. Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. NeuroImage 2016;129:460-69
4. Roy S, Butman JA, Pham DL, Initiative ADN. Robust skull stripping using multiple MR image contrasts insensitive to pathology. NeuroImage 2017;146:132-47
5. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54(3):2033-44
6. Salehi SSM, Erdogmus D, Gholipour A. Auto-context convolutional neural network (auto-net) for brain extraction in magnetic resonance imaging. IEEE Transactions on Medical Imaging 2017
7. Eskildsen SF, Coupé P, Fonov V, et al. BEaST: brain extraction based on nonlocal segmentation technique. NeuroImage 2012;59(3):2362-73
8. Beare R, Chen J, Adamson CL, et al. Brain extraction using the watershed transform from markers. Frontiers in neuroinformatics 2013;7
9. Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol. Skr. 1948;5:1-34
10. Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE transactions on medical imaging 2004;23(7):903-21
11. Souza R, Lucena O, Garrafa J, et al. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage 2017
Figures