3412
Surface Uniform Random Partition for Atlas-free Brain Network Analysis1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Shatin, Hong Kong, 2Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Shatin, Hong Kong, 3Philips Healthcare, Hong Kong, Hong Kong, 4Research Center for Medical Image Computing, Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Shatin, Hong Kong
Synopsis
Random partition is the cornerstone of atlas-free brain network analysis which can be used for multiscale analysis and comparison of cohorts with different brain sizes. The random parcels should be uniform to avoid additional variability from different parcel areas. In this study a uniform random partition of meshed surface is proposed considering geodesic distances and parcel areas. The partition results showed that proposed method can partition surface into any given number of parcels with similar areas. With repeating network analysis using proposed uniform parcels, results showed low intra-subject variations of global network measures.
Purpose
This study aims to partition brain gray matter and white matter interface surfaces into compact and uniform parcels for atlas-free brain network analysis. Compared with traditional atlas-based network analysis, the atlas-free analysis is unconstrained by pre-defined template and atlas, and thus it has been applied for multiscale analysis and comparison of cohorts with different brain sizes1–3. The nodes of atlas-free networks should be uniform parcels with similar areas or sizes to avoid introducing additional variability in charactering brain networks.Method
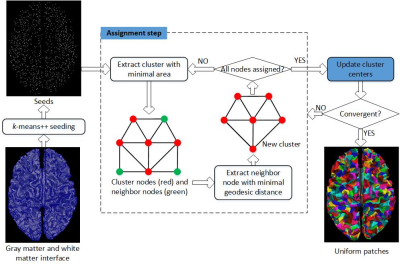
Proposed uniform random parcellation is depending on a graph clustering method using geodesic distances and parcel areas as shown in figure 1. Because shape of gray matter and white matter interface is very unconvex and complex, Euclidean distance is not appropriate for surface partition. For example, Euclidean distance between two points located at opposite side of sulcus is much shorter than the actual length of shortest curve connecting them. Therefore, geodesic distance, which is the length of shortest path between nodes, were employed in this study.
The gray matter and white matter interface surface was first constructed using FreeSurfer pipeline4. And then a weighted graph was constructed from the triangle mesh of interface surface by converting mesh vertices as graph nodes, and building edges between nodes within same triangles. Edge weights were set as spatial distances between vertices, and node weights were assigned as 1/3 sum of areas of triangles containing corresponding vertices.
To avoid sometimes poor partition from random initiation, a modified k-means++ algorithm was used for choosing initial seeds (i.e. cluster centers)5. The first seed was chosen randomly from all nodes followed by calculating geodesic distances between all the other nodes and the first seed using Dijkstra’s algorithm6. Then the next seed was chosen from nodes with top 5% largest geodesic distances to existing seeds, in order to ensure that new selected seed was always far away from existing seeds. The seeds except the first one were chosen according to a probability distribution with probability proportional to squared geodesic distances. Each time after choosing a new seed, the Dijkstra’s algorithm was recalled from new seed to update geodesic distances.
Similar to k-means clustering and simple linear iterative clustering7, the proposed graph clustering method is a two-step iterative optimization with an assignment step and an update step. In the assignment step, a cluster with minimal area and at least one unassigned neighbor node was first extracted. Then unassigned neighbor node with minimal tentative geodesic distance to the cluster center was allocated to this cluster. In the update step, a new cluster center was found using gradient descent search to minimize sum of geodesic distances between all cluster members and the new cluster center. The gradient descent search compared sums of geodesics distances from the old cluster center and its neighbors, and moved to the next node with minimal sum, and so on. The assignment-update loop was terminated until all seeds remained unchanged or iteration number reached a predefined limit.
Result
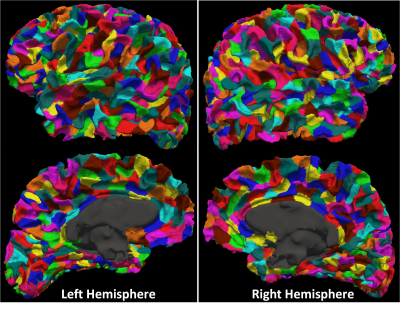
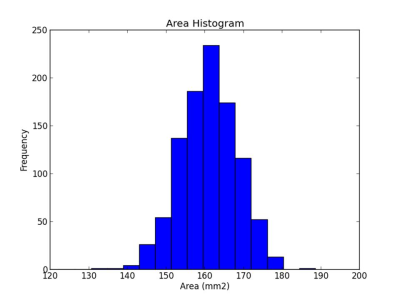
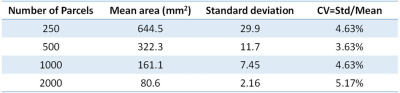
The gray matter and white matter interface surface was segmented into 250, 500, 1000, and 2000 parcels. Figure 2 and figure 3 shows the partition result and the histogram of parcel areas with 1,000 parcels respectively. Table 1 shows the coefficients of variation (CV), i.e. ratio of standard deviation to the mean, of parcel areas with different numbers of parcels.
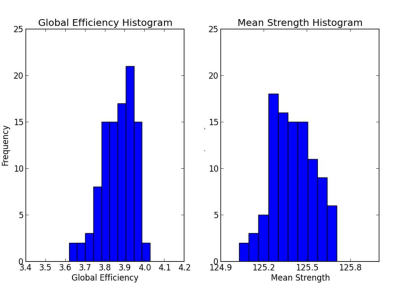
Structural networks were constructed from 1,000 parcels using counts of connecting fibers as edge weights by DSI-studio 100 times8. Global network measures including global efficiency and mean strength were calculated as shown in figure 3. The CV of global efficiency and mean strength are 2.11% and 0.12% respectively.
Conclusion
In this study, a uniform random partition method was proposed to partition gray matter and white matter interface surfaces into any given number of parcels with similar areas. The CV of proposed parcels’ areas was around 4% - 5%, much less than results of previous works, e.g. 52% (Echtermeyer, 813 parcels). And the repeated network analyses showed a low intra-subject variation of global network measures of atlas-free network analysis using proposed random parcels. Proposed method could benefit future brain structural and functional network analysis.Acknowledgements
The work described in this paper was supported by a grant from Research Grants Council of the Hong Kong Special Administrative Region, China (Project No.: CUHK 14113214), and a grant from the Innovation and Technology Commission (Project No.: GHP/028/14SZ, ITS/293/14FP).References
1. de Reus MA, van den Heuvel MP. The parcellation-based connectome: Limitations and extensions. Neuroimage. 2013;80:397-404. doi:10.1016/j.neuroimage.2013.03.053.
2. Tymofiyeva O, Ziv E, Barkovich AJ, Hess CP, Xu D. Brain without anatomy: Construction and comparison of fully network-driven structural MRI connectomes. Hayasaka S, ed. PLoS One. 2014;9(5):e96196. doi:10.1371/journal.pone.0096196.
3. Tymofiyeva O, Hess CP, Ziv E, et al. Towards the “baby connectome”: Mapping the structural connectivity of the newborn brain. Ben-Jacob E, ed. PLoS One. 2012;7(2):e31029. doi:10.1371/journal.pone.0031029.
4. Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781. doi:10.1016/j.neuroimage.2012.01.021.
5. Taghi M, Howard S. k-means++: the advantages of careful seeding. In: Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms. Association for Computing Machinery; 2007:1242. doi:10.1137/1.9781611973402.
6. Dijkstra EW, W. E. A note on two problems in connexion with graphs. Numer Math. 1959;1(1):269-271. doi:10.1007/BF01386390.
7. Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans Pattern Anal Mach Intell. 2012;34(11):2274-2282. doi:10.1109/TPAMI.2012.120.
8. Yeh F-C, Verstynen TD, Wang Y, Fernández-Miranda JC, Tseng W-YI. Deterministic Diffusion Fiber Tracking Improved by Quantitative Anisotropy. PLoS One. 2013;8(11):e80713. doi:10.1371/journal.pone.0080713.