3408
Sharing Radial GRAPPA Weight Sets Across k-Space to Decrease Memory Requirements for Real-Time Imaging1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States
Synopsis
This study examines the feasibility of reconstructing multiple neighboring k-space points from a single non-Cartesian GRAPPA weight set. This approach reduces both the time to calibrate the GRAPPA weights and the memory needed to store the weights with minimal loss of image quality in the reconstructed images.
Introduction
Through-time radial GRAPPA1 is a parallel imaging method that enables high frame rate visualizations and is well-suited to real-time, online image reconstruction2,3. However, a different set of GRAPPA weights is required to estimate each unacquired k-space point, resulting in long computation times. Furthermore, new GRAPPA weights must be calculated for each slice position and orientation. If these GRAPPA weights must be stored, a necessity for online reconstructions, this also results in large memory requirements. It would be beneficial to reduce the number of GRAPPA weights in order to shorten the calibration time and reduce latencies in image display. A smaller number of weights would also enable weight sets for several anticipated slice positions and orientations to be calculated and stored prior to accelerated data acquisition, allowing more flexible real-time imaging. This investigation examines the possibility of reusing GRAPPA weights for adjacent undersampled k-space points, decreasing the total number of weights required to reconstruct undersampled data for real-time imaging applications.Methods
Cardiac scans were performed on a 3T Skyra MRI (Siemens Healthineers, Erlangen, Germany) in short-axis orientation with a 30-channel cardiac array coil from a healthy volunteer in this IRB-approved study. Radial data were acquired with a balanced-SSFP sequence with the following parameters: FOV 300mm2, 128x128 matrix, 2.34x2.34mm2 resolution, 8mm slice thickness, 37° flip angle, and TR/TE=2.94/1.48ms. A calibration scan was acquired with 80 fully-sampled frames containing 144 radial projections. Real-time, undersampled data were acquired at three acceleration factors: R=6, 9, and 12 with 24, 16, and 12 projections per frame, respectively. All data were acquired during free-breathing without ECG gating.
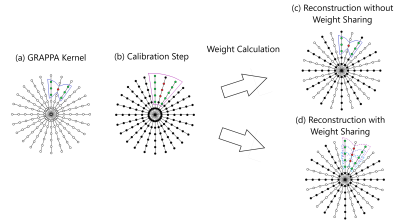
Radial GRAPPA weights were calibrated using 80 repetitions and a segment size of 4 (readout) x 1 (projections). In the proposed through-time radial GRAPPA reconstruction, instead of calculating a unique weight set for every target point, a single weight set was employed to reconstruct multiple adjacent points in the readout direction (Figure 1). The number of adjacent points that shared a single weight set was varied from 1 to 20, and the resulting images were compared to a reconstruction with no weight sharing (i.e. each missing point was generated with a unique weight set) using RMSE averaged over the entire image. All reconstructions were performed using a GPU-accelerated version of through-time radial GRAPPA in the Gadgetron framework2,4.
Results
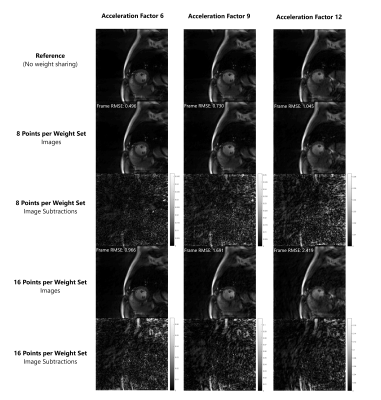
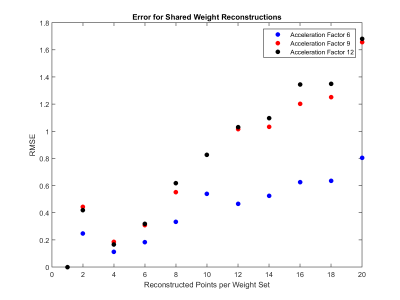
Sample images collected at acceleration factors of 6, 9, and 12 and reconstructed with two different levels of weight sharing are shown in Figure 2. There is little visual deterioration in image quality when 8 adjacent points share a weight set, but artifacts become visible when weight sharing is used across 16 points, particularly at higher acceleration factors. Overall, RMSE increases as the number of points reconstructed with a single weight set increases (Figure 3), but this error is relatively small for weight-sharing levels below 8 points.
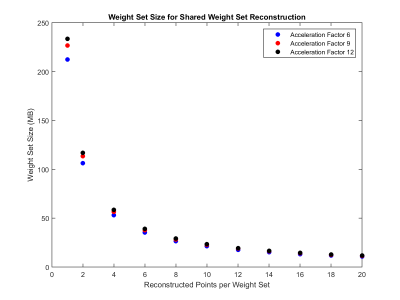
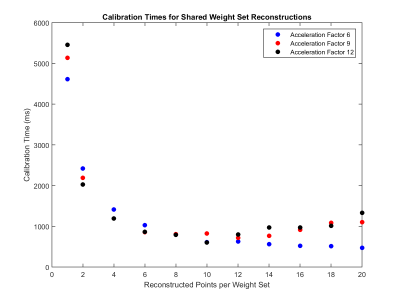
The total weight set size currently used in radial GRAPPA reconstructions is 212.3 MB for data collected at an acceleration factor of 6. As an example, when weight sets are shared among 8 points, they take up only 26.5 MB (Figure 4). This allows 8 times more slices or orientations to be stored and available to reconstruct images in different positions in real-time. There was also a decrease in the time required to generate the GRAPPA weights. Calculating a set of weights for each point took 4.62 seconds; when the weights were shared between 8 points, this process took only 1.03 seconds (Figure 5). Similar trends are seen for acceleration factors of 9 and 12.
Discussion
As the main advantage of radial GRAPPA is the ability to collect and reconstruct images in real-time, a prime application of this technique could be interventional MRI. This study found that sharing weight sets among neighboring points significantly decreases both the memory requirements for storing the weight sets, and the time required for calibration, while having little impact on the visual quality of the images. This allows more weight sets to be stored on the reconstruction computer, and gives more freedom to change slices or orientations during real-time imaging for interventional procedures. While the image RMSE does increase with the number of points reconstructed per weight set, these increases are relatively small.Conclusion
These results show that weight set sharing in through-time radial GRAPPA can be used as an effective method of both decreasing memory requirements and calibration times while preserving image quality. This would allow more weight sets to be stored before beginning an interventional procedure, thus allowing allow more flexibility to change slices or orientations during a procedure for better views.Acknowledgements
Siemens Healthcare, R01EB018108, R01DK098503, and R01HL094557.References
1. Seiberlich, Nicole, et al. Improved Radial GRAPPA Calibration for Real-Time Free-Breathing Cardiac Imaging. Magn. Reson Med. 2011 65:492-505
2. Franson D, Ahad J, Hamilton J, Lo W, Jiang Y, Chen Y, Seiberlich N. Real-time 3D cardiac MRI using through-time radial GRAPPA and GPU-enabled reconstruction pipelines in the Gadgetron framework. In: Proc. Intl. Soc. Mag. Reson. Med. 25.; 2017. p. 448.
3. Saybasili, Haris, et al. Real-time imaging with Radial GRAPPA: Implementation on a heterogeneous architecture for low-latency reconstructions. Magn. Reson Med. 2014 32(6):747-58.
4. Hansen MS, Sørensen TS. Gadgetron: An open source framework for medical image reconstruction. Magn. Reson. Med. 2013;69:1768–1776. doi: 10.1002/mrm.24389.
Figures