3404
Translating the Human Connectome Project to Marmoset Imaging: 16-Channel Multi-Array Coil and HCP-Style MRI Protocols and PreprocessingYuki Hori1, Joonas Autio1, Masahiro Ohno1, Yoshihiko Kawabata2, Yuta Urushibata3, Katsutoshi Murata3, Masataka Yamaguchi1, Akihiro Kawasaki1, Chiho Takeda1, Chihiro Yokoyama1, Matthew F Glasser4,5, and Takuya Hayashi1
1Center for Life Science Technologies, RIKEN, Kobe, Japan, 2Takashima Seisakusho Co. Ltd., Hino, Kiribati, 3Siemens Healthcare Japan, Tokyo, Japan, 4Department of Anatomy and Neurobiology, Washington University in St. Louis, St. Louis, MO, United States, 5St. Luke's Hospital, St. Louis, MO, United States
Synopsis
The common marmoset is increasingly used as a non-human primate model to understand the organization of the brain. Better cross species comparisons can be achieved by adapting methods from the Human Connectome Project. Here, we show a customized 16-channel receiver coil designed for the marmoset brain and present the initial imaging results on a 3T MRI scanner with powerful gradients. The coil had high signal-to-noise ratio and B1 transmit homogeneity. In-vivo marmoset data, acquired and preprocessed using HCP-style methods, provided high-resolution images, allowing cortical mapping of myelin, thickness, and structural and functional connectivity, enabling high quality cross-species comparisons.
INTRODUCTION
The common marmoset is increasingly used as a non-human primate model to explore cortical organization underlying behaviors and disorders. Advances in genetic/viral techniques also attract researchers to use this species [1]. The human Connectome Project (HCP) has revealed that high-resolution multi-modal MRI provides powerful means to address cortical architecture and connectomics in living humans [2, 3]. Applying corresponding approaches will produce better results for comparative neuroscience, but this is challenging because of the cross-species diversity in brain size, shape and cortical convolutions. Past marmoset MRI studies have used dedicated high-field animal scanners, rather than a high performance clinical system like the HCP, nor have they generally used multi-channel head coils and HCP-Style sequences. Here, we show the result of our efforts to develop a 16-channel multi-array RF receive coil for marmoset brain imaging for a 3T-MRI scanner with powerful gradients. Basic quality assessments and adaption of HCP-style data acquisition and preprocessing pipelines are presented.METHODS
The coil frame geometry was designed to closely fit the largest head dimension in our marmoset MRI database (Fig. 1A). The locations and overlaps of 16 coil elements were designed using a 3D design software (Fig. 1B) and drawn on the surface of the coil frame (Fig. 1C). Each element was composed of a circular thin wire of polyester coaxial copper cable, placed overlapping with each other to reduce coupling (Fig. 1D). The coil was designed for a 3T-MRI scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany), which has powerful gradients (80mT/m). Element coupling was assessed with noise correlation measurements. A custom phantom of a marmoset head was used to evaluate B1 field inhomogeneity. Four anesthetized common marmosets (male, 381±5.3 g, 5.7±4.0 years old) were used for in-vivo MRI scanning with same pulse sequence types as in HCP (e.g. MPRAGE, SPACE, multi-band EPI) and parameters designed in line with principles of HCP [4]. The structural T1w and T2w images were obtained with isotropic spatial resolution of 0.18mm. Two resting state fMRI (rfMRI) scans, with opposing phase direction, were obtained with TR/TE=0.76sec/30msec, scan time of 50min, and spatial resolution of 1.1 mm, which was derived by comparing histograms of cortical thickness between human and marmosets to be roughly equivalent to 2mm in the human. High-resolution dMRI (0.8 mm) was scanned in three shells with the highest at b= 3000 s/mm2 and 500 gradient directions in scan time of 30 min. All the data were analyzed and preprocessed with the HCP pipelines to correct for motion, distortion and intensity biasfield. Structural MRI was used for estimating a subcortical segmentation, cortical surface, and thickness and myelin maps. fMRI was analyzed with independent-component analysis to enable denoising and delineation of functional networks. dMRI was analyzed with a NODDI model for neurite imaging [5], bedpost fiber orientation modeling and probabilistic tractography [6].RESULTS
The noise correlation coefficients (Fig. 2A) ranged from 0.01 to 0.67 with an average value of 0.29 ± 0.18. The B1 transmit field was quite homogeneous across brain regions (91.3 ± 1.5 [deg]), and the B1 receive map showed higher signal sensitivity in the periphery than in the center of the phantom (Fig. 2C, 2D) as expected. High-resolution structural T1w and T2w images in the marmosets provided high SNR and HCP-style biasfield correction achieved good homogeneity and contrast between grey and white matter (Fig. 3A, 3B), which further allowed us to obtain an accurate subcortical segmentation, surface estimation and myelin and thickness maps (Fig. 3C, 3D). The rfMRI data had higher temporal SNR than the HCP data and was corrected for motion and distortion and structured noise was removed using ICA+FIX [7]. Independent components representing several neural functional networks were found (Fig. 3E). Diffusion tractography successfully captured a known connection between the insula and anterior cingulate cortex (ACC), which is challenging to track because of crossing and kissing of longitudinal fascicules, commissural fibers and corona radiata (Fig. 3F).DISCUSSION
Our custom marmoset-dedicated 16-channel coil, implemented for a high-gradient 3T MRI scanner, provided images with excellent SNR and homogeneity, and allowed us to adapt HCP-style high-resolution image acquisitions needed for mapping the marmoset connectome. HCP-style preprocessing generated maps of myelin, cortical thickness, and cortical neurite indices that agreed with those in the previous reports in macaques and humans. Functional and structural connectivity analyses also provided high quality results including mapping connections that have previously been seen with the HCP data in humans.CONCLUSIONS
Our strategy of combining multi-array RF coil and high-gradient 3T scanner is potentially useful to establish marmoset connectomics in harmonized manner with human connectome.Acknowledgements
We thank Prof. Van Essen DC in Washington University School of Medicine. This research is partially supported by the program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from Japan Agency for Medical Research and development, AMED, by MEXT KAKENHI Grant (16H03300,16H03306, 16H01626, 15K12779), and by a grant from Sumitomo Dainippon Pharma Co., Ltd.References
[1] Okano et al. (2016) Neuron 92, 582-590 [2] Glasser, M.F. et al. (2013) NeuroImage 80, 105–124 [3] Van Essen, D.C. et al. (2012) NeuroImage 62, 2222–2231 [4] Glasser, M.F. et al. (2016) Nat. Neurosci. 19, 1175–1187 [5] Zhang, H. et al. (2012) NeuroImage 61, 1000–1016 [6] Behrens T.E.J. et al. (2007) NeuroImage 34, 144-155 [7] Griffanti et al. (2014) NeuroImage 95, 232-247Figures

Figure 1. The
development of marmoset 16-channel receive coil. Coil frame (A) and
element locations (B) were designed using 3D digital design software.
Element locations were drawn on the coil frame (C), and component parts
were symmetrically placed on the surface (D).
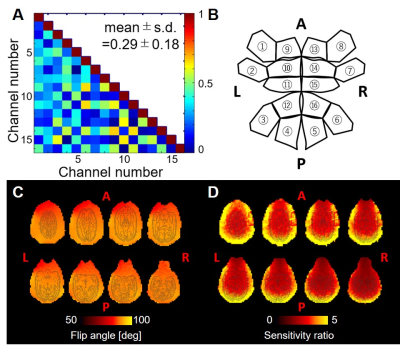
Figure 2. Characteristics
of the marmoset coil. (A) Noise correlation matrix among each channel
and (B) corresponding channel number in expansion diagram of the
marmoset coil. Mean and standard deviation of correlation coefficient are
0.29±0.18. (C) B1 transmit flip angle maps of the marmoset
phantom were acquired using 90 degrees target flip angle. (D) B1
receive maps of the marmoset phantom were calculated by images using marmoset
coil divided by those using human body coil.
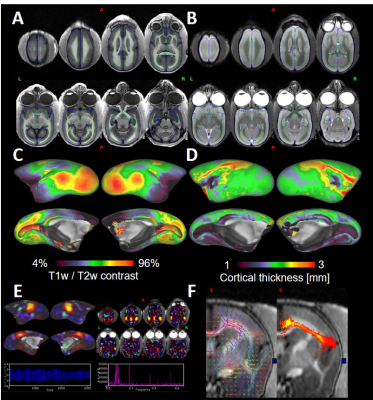
Figure 3. Representative
T1w (A) and T2w (B) images. Green and blue lines were white and pial surfaces,
respectively. Myelin (C) and cortical thickness (D) maps were presented on the
inflated cortical surface. (E) Default-mode
network (cortical surface, volume images, power spectral density and
time-course). (F) Probabilistic
tractography result between anterior cingulate and insular cortex.