3401
2D selective excitation with UNFOLD for 4D Flow Imaging1Physikalisch-Technische Bundesanstalt (PTB), Braunschweig and Berlin, Germany
Synopsis
4D flow MRI allows to quantify the velocity vector field non-invasively in-vivo. However, it still suffers from long acquisition times and low temporal and spatial resolution. Here, we accelerate acquisition time and increase temporal resolution without loss of spatial resolution by combining 2D selective excitation and UNFOLD. 2D selective excitation allows to limit the field-of-view in phase encoding direction and thus acquisition time, whereas UNFOLD grants to increase temporal resolution.
Introduction
4D flow MRI1 allows to quantify the velocity vector field non-invasively in-vivo. However, long scan time and low temporal or spatial resolution are still challenges of the technique. In this work, we aim to address long scan time and low temporal resolution by two complementary methods. First, we excite 2D selectively a reduced field-of-excitation (FOX) enabling a reduced field-of-view (FOV) in phase encoding (PE) direction (FOVPE). However, the long duration of 2D RF-pulses leads to long TR and thus reduced temporal resolution. Therefore, we additionally apply UNFOLD2,3, an acceleration technique for dynamic imaging that has been applied successfully to 4D flow4, to double temporal resolution.
Methods
2D Selective Excitation with 4D Flow MRI
2D selective RF-pulses with spiral k-space trajectory (Fig.1a) and bandwidth-time-product $$$BWTP=1.2$$$ are designed in the small flip angle regime according to Pauly5 to excite a $$$(6\times6\times\infty)\,\text{cm}^3$$$ FOX. The trajectory starts at $$$k_\text{max}=2.51\,\frac{\text{rad}}{\text{cm}}$$$ and spirals inwards with $$$n=10$$$ turns in $$$T=3.5\,\text{ms}$$$. The actual gradient waveform was measured6 and integrated into the RF-pulse calculation. Fig.1b shows the resulting excitation profile.
The 2D excitation pulse was integrated into a Cartesian GRE ECG-triggered 4D flow MR sequence1.
In-Vivo Experiment
A volunteer was scanned according to an approved ethical protocol and with informed consent at 3T (Magnetom Verio, Siemens, Germany). 4D flow imaging was performed using two protocols with varying number of PE-lines and excitation pulses to acquire two data sets: I) conventional 1D-slab-selective excitation and 96 PE-lines (full FOVPE); II) 2D selective excitation and 48 PE-lines (FOVPE/2). For all scans we acquired two PE-lines and four velocity encodings per cardiac phase, resulting in a temporal resolution $$$\Delta\,t=8\,T_\text{TR}$$$. Note that the different RF-pulse durations (Tab.1) result in different temporal resolutions, $$$\Delta\,t^\text{1D}=43.2\,\text{ms}$$$ and $$$\Delta\,t^\text{2D}=64.8\,\text{ms}$$$. Acquisition parameters are listed in Tab.1.
Reconstruction with UNFOLD
The sequence acquisition order (two lines per cardiac phase) allows to split odd and even lines of each cardiac phase into two separate time frames. Thereby, time-resolution is doubled, FOVPE is halved and the set of phase-encodes is shifted by half a line from time frame to time frame (Fig.2a). The obtained complex datasets were Fourier transformed to complex image space, where they show undersampling artefacts (Fig.2b). Subsequently, a fast Fourier transform (FFT) was applied along the temporal dimension. Undersampling artefacts were then removed with a Fermi-filter $$$f(E)=\left (1+\exp{\left(\frac{|E|-E_\text{f}}{k_\text{B}T}\right)}\right)^{-1}$$$ in frequency space, where $$$E_\text{f}=0.5N_y,\,k_\text{B}T=0.022N_y$$$ and $$$N_y$$$ the Nyquist frequency (Fig.2c). UNFOLDed images are finally obtained by applying an inverse FFT to the filtered temporal frequencies (Fig2d).
UNFOLD was applied to
in-vivo data set II) as explained above. Furthermore, data set I) was
retrospectively four-fold undersampled in PE-direction (data set III) and reconstructed
with and without UNFOLD.
Results
Fig.3a shows magnitudes and absolute velocities in the middle cerebral arteries during systole ($$$140.4$$$ and $$$145.8\,\text{ms}$$$ after ECG-trigger) for both data sets. Besides minor aliasing of residual sub-cranial fat signal outside the FOX, successful 2D selection and flow quantification was obtained. Scan times and temporal resolutions are listed in Fig.3a.
Fig.3b shows the images resulting from data set II) split into twice the number of cardiac phases and reconstructed with/without UNFOLD (columns 3&4). UNFOLD effectively removed the aliasing artifacts seen in column 3, resulting in image quality equivalent to Fig.3a (right), but with doubled temporal resolution. For comparison, applying UNFOLD to dataset III) does not fully remove all aliasing artifacts (column 1&2).
The benefit of applying
UNFOLD to 2D selective excitation with reduced FOV to improve the temporal
resolution is shown in Fig.4 with time-resolved flow quantification through the internal
carotids and basilar artery (Fig.4b). Due to the lower temporal resolution of
data set II ($$$\Delta\,t^{II}=64.8\,\text{ms}>\Delta\,t^{I}=43.2\,\text{ms}$$$),
the peak volume flow is underestimated by $$$\frac{\Delta\hat{v}}{\hat{v}}=11.37\%$$$
compared to data set I. The improved temporal resolution of data set II
combined with UNFOLD ($$$\Delta\,t=32.4\,\text{ms}$$$) (Fig.4a) allows to
correctly capture peak volume flow ($$$\frac{\Delta\hat{v}}{\hat{v}}=3.63\%$$$)
while reducing the scan time by $$$50\,\%$$$.
Discussion and Conclusion
In this work we combine 2D RF excitation with UNFOLD reconstruction to allow for reduced FOV imaging with increased temporal resolution. A main challenge of 2D selective RF excitation is the longer RF pulse duration leading to longer TR and thus to impaired temporal resolution counteracting the benefits of FOX/FOV reduction. UNFOLD was successfully applied to address this issue leading to improved quantification in 4D flow MRI. To further accelerate 2D excitation we aim to apply parallel transmission strategies, which will then also reduce the residual fat artifact that is apparent in the images.
Acknowledgements
References
1. Markl M et al. 4D flow MRI, J. Magn. Reson. Imaging 2012;36:1015-1036.
2. Madore B et al. Unaliasing by Fourier-Encoding the Overlaps Using the Temporal Dimension (UNFOLD), Applied to Cardiac Imaging and fMRI. Magn Reson Med 1999;42:813-82.
3. Zhao L et al. Reduced Field-of-View MRI With Two-Dimensional Spatially-Selective RF Excitation and UNFOLD. Magn Reson Med 2005;53:1118-112.
4. Braig M et al, Evaluation of Accelerated Preclinical 4D-Flow Imaging with UNFOLD, ISMRM Conference Proceedings 2017.
5. Pauly J et al. A k-Space Analysis of Small-Tip-Angle Excitation. J. Magn. Reson. Imaging 1989;81:43-56.
6. Duyn J H et al. Simple Correction Method for k-Space Trajectory Deviations in MRI. J. Magn. Reson. 1998;132:150-153.
Figures
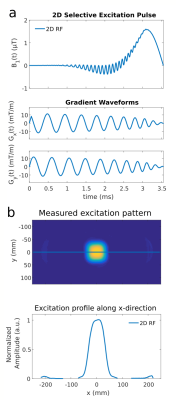
Figure 1
(a) RF and gradient diagram of the spiral k-space trajectory of our 2D RF-pulse
(b) Resulting measured excitation profile of the 2D RF-pulse.
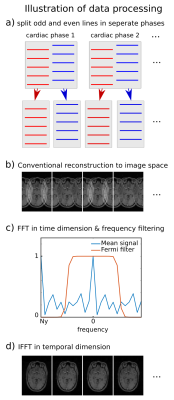
Figure 2
Illustration of data processing with UNFOLD
(a) Data of sequence which acquired 2 lines per phase is split into separate time frames. The set of k-space lines is now shifted by half a line between adjacent frames.
(b) Each frame is conventionally reconstructed to complex image space, showing aliasing artefacts, which change sign at the Nyquist frequency Ny.
(c) A FFT is applied in time dimension and the resulting signal is filtered with a Fermi filter to remove aliasing artefacts.
(d) The IFFT in the temporal dimension produces unaliased dynamic images with doubled temporal resolution.
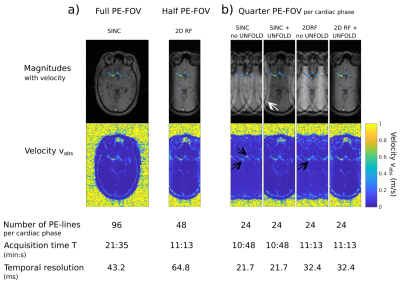
Figure 3
GRE magnitude (gray-scale) and velocity images (color-coded).
(a) Full FOVPE (left) and FOVPE/2 (right) imaged with 1D- and 2D-selective excitation respectively showing the Circle-of-Willis during systole.
(b) Comparison of four different dynamic imaging methods using a quarter of PE-k-space-lines: i) conventional 1D-slab-selective excitation, two-fold aliasing is visible in magnitude and velocity (arrows); ii) with UNFOLD only one-fold aliasing is observed (arrow) iii) 2D RF without UNFOLD, two-fold aliasing is still visible (arrow); iv) with 2D RF and UNFOLD aliasing is comparable to (a,left).
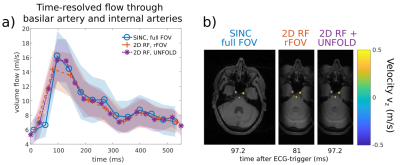
Figure 4
(a) Comparison of time-resolved flow through the basilar artery and internal carotids between i) 1D-selective excitation with full FOV, ii) 2D-selective excitation with half a PE-FOV, and iii) 2D-selective excitation combined with UNFOLD to double time resolution. The errorbounds are the standard deviation of all 11 voxels of the basilar artery and internal carotids.
(b) GRE Magnitudes (gray-scale) and velocities in z-direction (color-coded) of the slice in which flow is analysed in (a).
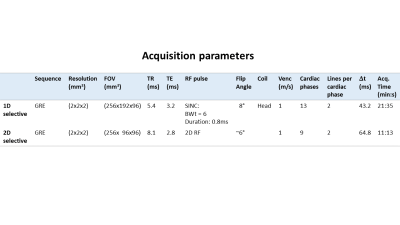
Table 1
Acquisition parameters.
For the 2D RF-pulse the given flip angle is a nominal angle. The given acquisition time is nominal for an acquisition window of i) 562ms and ii) 584ms.