3384
Single Point Dixon Reconstruction of Whole-Body Scans Using a Convolutional Neural Network1Department of Radiology, Uppsala University, Uppsala, Sweden, 2Antaros Medical, BioVenture Hub, Mölndal, Sweden
Synopsis
Reconstructions of water and fat images are clinically useful for removing obscuring fat signal. It can also be useful in for example obesity related research, measuring for example different adipose depot volumes. Normally reconstructions would be performed using at least two echos, which requires about twice as much time as collecting a single echo. Therefore, using only a single echo would reduce the required scan time drastically. In this abstract a method for reconstruction of water and fat images from a single echo is introduced, using a convolutional neural network. We conclude from visual evaluation that the results are promising.
Introduction
The water and the fat signal of spoiled gradient echos are separable. The performance of the separation is known as Dixon reconstruction. The resulting water images are clinically useful, where fat is often obscuring, and the fat images can be useful in for example obesity related research. Typically multiple echoes, also referred to as points, are used, since the single-point Dixon reconstruction is underdetermined. Considering a single voxel only, it is impossible to disentangle the water and the fat signal, due to B0-inhomogeneities. However, using additional knowledge, such as spatial awareness and the knowledge of spatial smoothness of the B0-inhomogeneities makes separation possible. This has previously been performed using region-growing algorithms.1,2 However, these methods are not widely used.
Previously neural networks have been used for MRI reconstructions, mostly for reconstructions of undersampled k-space data to image-space data,3,4 but also for refinement of water-fat reconstructions5 and in magnetic resonance fingerprinting.6
Methods
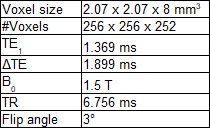
Imaging data came from the POEM-study.7 Whole-body scans of 240 subjects, all at age 50, were included. Three monopolar echoes were acquired. Further protocol information can be found in table 1.
The U-net is a convolutional neural network originally used for semantic segmentation of biomedical images.8 The network was implemented in TensorFlow9 and modified in the following ways: As input the real and imaginary parts of the first echo was used. Padding was performed inside the network after the convolutions to make the output the same size as the input. Only one feature map was used as output, corresponding to the fat fraction. Reconstructions using the method analytical graph cut (AGC)10 using all echoes were performed and the resulting fat fractions were used as reference during training. As cost function voxelwise squared difference between the reference and the output was used. Background voxels were ignored in the cost function. A GeForce GTX 1060 with 3 GB of memory was used for training. For reconstruction both the GPU and an Intel Core i5-6300HQ CPU were used.
The scans were randomly assigned into training and validation sets, with 70% in the training set.
The Adam optimizer11 was used. The following parameters were used: Initial learning rate: 0.001, β1 = 0.9, β2 = 0.999, ε = 1e-8. From the training set 200 random axial slices were chosen and used to train the network one at a time. After this the learning rate was decayed by 5%. This process was iterated 100 times.
Computation time for training with the GPU was noted. For reconstruction both the computation time using the GPU and the CPU were noted.
After training, fat fraction images were calculated for all subjects in the validation set. Water and fat signal images were calculated from the resulting fat fraction images and the absolute value of the first echo by using a multi-fat-peak signal model.12
Quality of images were determined by visual inspection. Axial slices of fat fraction images of both the reference and reconstructed using the proposed method were scrolled through jointly.
Results
Training took 58 minutes. Reconstruction took 0.034s per slice, equal to 8.6s per whole body scan, using the GPU, and 1.9s per slice, equal to 7.9 minutes per whole body scan, using the CPU.
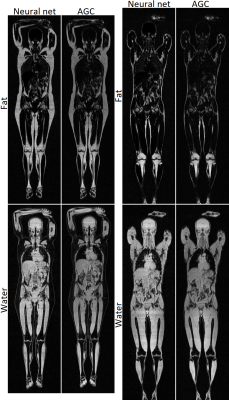
Two example reconstructions are shown in Figure 1.
Only a few subjects had a fatty liver. In these cases the network failed to identify this, and instead calculated a normal liver fat content. Other than this no systematic or major errors were found in the reconstructions performed using the network. The errors that were noted were on par with those found in the reference reconstructions.
Discussion
While the reconstructed images are generally of good quality, there are some persisting errors in the reconstruction, mainly an inability to properly calculate the liver fat content.
Modification of the network to suit this particular problem better could potentially resolve this. For example a 3D architecture could be attempted, or a longer training period could be used. Acquiring more training data, especially of patients with an elevated liver fat content, could potentially resolve the problems.
The measured reconstruction times indicate that the method is feasible to use even without a GPU.
Conclusion
The results of the proposed method indicate that conventional two point Dixon acquisition could potentially be replaced with a single point acquisition in several applications, thereby significantly reducing the required scan time.
This would be especially useful in situations that requires fast scanning, such as when using dynamic contrast, or having subjects that have trouble keeping still, e.g. children. It could also be useful for quicker attenuation scans in PET/MR-scanners.
Acknowledgements
Swedish Research Council (2016-01040)References
1. Ma J. Breath-hold water and fat imaging using a dual-echo two-point Dixon technique with an efficient and robust phase-correction algorithm. Magn Reson Med 2004; 52: 415–419.
2. Berglund J, Ahlström H, Johansson L, Kullberg J. Single-image water/fat separation. In: Proceedings of the 18th Annual Scientific Meeting of the ISMRM, Stockholm, Sweden, 2010. (abstract 2907).
3. Hammernik K. Insights into Learning-Based MRI Reconstruction. In: Educational Course during the 24th Annual Scientific Meeting of the ISMRM, Honolulu, USA, 2017.
4. Knoll F. Leveraging the Potential of Neural Networks for Image Reconstruction. In: Educational Course during the 24th Annual Scientific Meeting of the ISMRM, Honolulu, USA, 2017.
5. Gong E, Zaharchuk G, Pauly J. Improved Multi-echo Water-fat Separation Using Deep Learning. In: Proceedings of the 24th Annual Scientific Meeting of the ISMRM, Honolulu, USA, 2017. (abstract 5657).
6. Cohen O, Zhu B, Rosen MS. Deep learning for fast MR Fingerprinting Reconstruction. In: Proceedings of the 24th Annual Scientific Meeting of the ISMRM, Honolulu, USA, 2017. (abstract 0688).
7. http://www.medsci.uu.se/poem
8. Ronneberger O, Fischer P, Brox T. (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J., Wells W., Frangi A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham
9. Abadi, M, Agarwal A, Barham P et al. (2015). TensorFlow: Large-scale machine learning on heterogeneous systems. http://download.tensorflow.org/paper/whitepaper2015.pdf.
10. Andersson J, Malmberg F, Ahlström H, Kullberg J. Analytical Three-Point Dixon Method Using a Global Graph Cut. In: Proceedings of the 24th Annual Scientific Meeting of the ISMRM, Singapore, Singapore, 2016. (abstract 3278).
11. Kingma DP, Ba JL. Adam: A Method for Stochastic Optimization. arXiv:1412.6980 [cs.LG], December 2014.
12. Yu H, Shimakawa A, McKenzie CA et al. Multiecho water-fat separation and simultaneous R2* image estimation with multifrequency fat spectrum modeling. Magn Reson Med 2008; 60: 1122–1134.