3383
A Machine Learning Approach for Mitigating Artifacts in Fetal Imaging due to an Undersampled HASTE Sequence1Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States, 2Boston Children’s Hospital, Boston, MA, United States, 3IMES, MIT, Harvard-MIT Health Sciences and Technology, Cambridge, MA, United States, 4Athinoula A. Martinos Center for Biomedical Imaging at Massachusetts General Hospital, Boston, MA, United States, 5Electrical Engineering and Computer Science, MIT CSAIL, Cambridge, MA, United States, 6Electrical Engineering and Computer Science, Institute for Medical Engineering and Science, MIT, Cambridge, MA, United States
Synopsis
This work investigates using deep learning to mitigate artifacts in fetal images resulting from accelerated acquisitions. We applied an existing deep learning framework to reconstruct undersampled HASTE images of the fetus. The deep learning architecture is a cascade of two convolutional neural networks combined with data consistency layers. Training and evaluation were performed on coil-combined and reconstructed HASTE images with retrospective undersampling. The datasets derived from imaging of ten pregnant subjects, GA 19-37 weeks, yielding 3994 HASTE slices. This approach mitigates artifacts from incoherent aliasing with residual reconstruction errors in high spatial frequency features in the phase encoding direction.
Introduction
T2-weighted fetal brain imaging is an important component of diagnostic MRI in pregnancy. Currently, single-shot HASTE imaging is the dominant means of collecting such contrast in the fetus due to its motion robustness, but tradeoffs due to the long refocusing train include reduced quality of contrast when compared to multi-shot turbo-spin echo acquisitions [1].
With the ultimate goal of improved T2 contrast in HASTE, we evaluate an image reconstruction of a reduced-length refocusing train in HASTE imaging. Undersampling phase encoding lines in HASTE results in aliasing and resolution tradeoffs, and this work investigates the potential to use deep learning to correct such artifacts in image reconstructions of retrospectively undersampled images. We train and test on HASTE fetal images using the deep learning framework in [3] and demonstrate mitigation of aliasing artifacts as well as the presence of residual errors in high spatial frequency features.
Methods
Based on the proposed architecture in [3], we implemented a cascade of two convolutional neural networks (CNNs) [2], each comprised of one convolution filter, with data consistency layers interleaved between the CNNs. The overall network takes a 256x256 single-channel, real-valued image and outputs a same-sized corrected image.
The network was trained and tested on retrospectively undersampled images. We applied a 2D-FFT to the images, and undersampled the resulting k-space data along the presumed phase encode axis (i.e., y-axis of the image) using a variable density Gaussian Cartesian mask (Fig. 1) and sampling the ACS lines. The variance of the Gaussian distribution was varied to simulate acceleration factors of approximately R=3 and R=6. The original images served as the ground truth reconstruction. Image similarity was measured using normalized RMSE.
The dataset consist of 10 pregnant mothers who ranged in gestational age from 19 to 37 weeks. The training set had 7 patients’ data of 2886 images and the test set had 3 patients’ data of 1108 images. Images were obtained from 10 previous clinical scans of pregnant mothers at Boston Children’s Hospital with institutional review board approval. The images were derived from k-space data obtained by vendor reconstructions of GRAPPA=2 HASTE with partial Fourier acquisition.
Results and Discussion
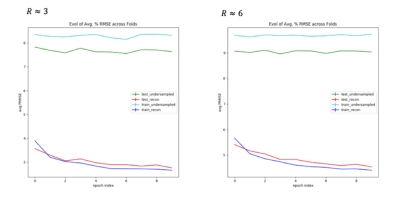
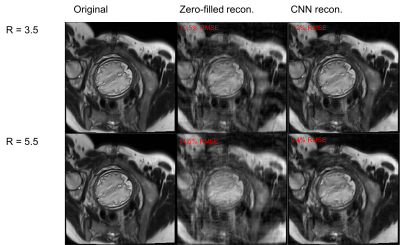
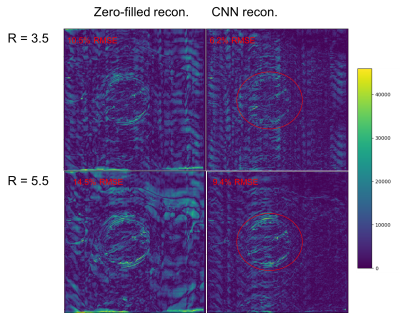
Fig. 2 shows metrics on the overall performance of the network and Fig. 3 and 4 show sample reconstruction results.
Compared with naïve zero-filled reconstruction of the retrospectively undersampled k-space data, the network reconstruction achieves 4-5% lower normalized RMSEs. The representative reconstructions shown in Fig. 3 and the corresponding error maps in Fig. 4 demonstrate that these improvements are mainly due to removal of incoherent artifacts but that the largest errors in the reconstructions correspond to high-spatial-frequency features in the simulated phase-encoding direction of the fetal anatomy.
To facilitate reconstruction over these anatomical details, we explored increasing sampling of higher spatial frequency content through a uniform density sampling mask. The resulting network reconstructions exhibited significantly higher normalized RMSEs (e.g., 45% for R=6) compared to reconstructions for the Gaussian distribution.
Conclusion
The potential for deep learning to mitigate undersampling artifacts motivates further work on prospective undersampling in HASTE and deep learning reconstruction to improve T2 contrast in fetal imaging. Our investigation using an existing deep learning framework shows that the network has higher normalized RMSEs for R=6 than R=3 and that network has higher residual error over high spatial frequency features for R=6 than R=3. To address these issues, future work includes the design of undersampling patterns that optimally trade off aliasing and resolution. We plan to investigate combining deep learning reconstruction with existing reconstruction approaches, including parallel imaging and compressed sensing. Further gains may derive from prioritizing reconstruction of some regions of interest (e.g. fetal brain) vs. others within the field of view of the image.Acknowledgements
NIBIB R01EB017337; NICHD U01HD087211; EECS MIT Quick Innovation Fellowship.References
1. Gholipour, Ali, et al. “Fetal MRI: A technical update with educational aspirations.” Concepts in Magnetic Resonance Part A, vol. 43, no. 6, 2014, pp.237-266., doi:10.1002/cmr.a.21321.
2. https://github.com/js3611/Deep-MRI-Reconstruction
3. Schlemper, J., Caballero, J., Hajnal, J., Price, A., Rueckert D.: A Deep Cascade of Convolutional Neural Networks for MR Image Reconstruction. arXiv preprint arXiv:1703.00555 (2017)
Figures