3375
MultiNet PyGRAPPA: A Novel Method for Highly Accelerated Metabolite Mapping1Max Planck Institute for Biological Cybernetics, Tuebingen, Germany, 2IMPRS for Cognitive and Systems Neuroscience, Eberhard-Karls University of Tuebingen, Tuebingen, Germany, 3Institute of Physics, Ernst-Moritz-Arndt University Greifswald, Greifswald, Germany
Synopsis
In this work, a novel acceleration method (MultiNet PyGrappa) is introduced which enables high in-plane acceleration factors for non-lipid suppressed 1H MRSI data. By using a variable density undersampling scheme and reconstructing the missing data points with multiple neural networks, this method enables a more robust reconstruction of highly undersampled data. High resolution metabolite maps acquired at 9.4T in the human brain using the proposed method are presented.
Introduction
The in-plane acceleration factors that can be achieved through parallel imaging for 1H MRSI are limited by the inherent low SNR of the MRSI data and the noise amplification resulting from higher acceleration factors1. In addition to that, for ultra-short TR and TE 1H MRSI without any lipid suppression, the problem is more severe2 since the strong lipid signals present in the data can deteriorate the metabolite maps when the reconstruction is unable to fully resolve all the aliasing.
In this work we introduce a novel and improved reconstruction method (MultiNet PyGRAPPA) by using multiple neural networks for reconstruction, and further combining that with a variable density undersampling scheme. Using this method, highly accelerated metabolite maps acquired at 9.4T are presented.
Methods
Five healthy volunteers were scanned using a Siemens 9.4T whole-body human MRI scanner with an in-house developed 18Tx/32Rx RF coil3. Fully-sampled MRSI data from a single slice were acquired from each volunteer using a non-lipid suppressed ultra-short TR and TE FID MRSI sequence4-6 with the following parameters: FOV = 200x200mm; slice thickness = 10mm; matrix size = 64x64; TE = 1.56ms; TR = 300ms; spectral bandwidth = 8000Hz; total scan time = 15 mins. After each MRSI scan, a high resolution reference anatomical image was acquired using a 2D FLASH sequence.
The fully sampled data was retrospectively undersampled by a factor of R=2x2. Two different GRAPPA reconstruction methods were compared: 1) a conventional GRAPPA7 reconstruction where the kernel was calibrated on the FLASH anatomical image, 2) a novel neural-network based GRAPPA reconstruction (MultiNet) where the kernel is obtained using neural networks. The weights of the network were trained on the scout image using the LBFGS algorithm and were used to reconstruct the missing points in the MRSI image.
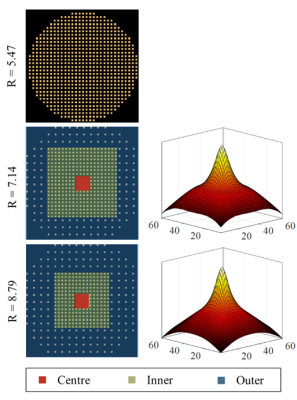
In addition to the fully sampled data, accelerated MRSI data were acquired from each volunteer using two variable density acceleration masks (pyGrappa) with R=7 and R=9 (Figure 1). The corresponding sampling density distributions are also shown in this figure. The resulting scan times were about 2.8 and 2 minutes for R=7 and 9, respectively.
The MultiNet reconstruction was modified and separated to different regions in k-space for reconstructing the variable density data. Multiple neural networks with different properties were trained for each k-space region. The hidden layer of the neural network was changed to be nonlinear for the reconstruction of the outer k-space region to account for the nonlinear relationship between the missing and acquired data points due to higher levels of noise present in the outer regions of k-space.
Results/Discussion
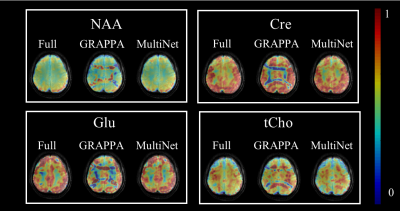
Figure 2 shows the metabolite maps of 4 major metabolites for the fully sampled data, conventional GRAPPA (R=2x2) and MultiNet (R=2x2). The advantage of the proposed MultiNet reconstruction over conventional GRAPPA can be seen from the residual aliasing artifacts present in the metabolite maps resulting from the conventional GRAPPA reconstruction.
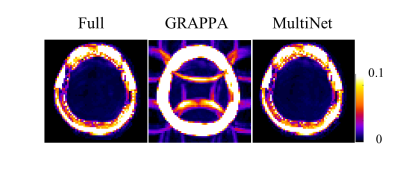
Figure 3 shows the lipid contamination maps for the different reconstruction methods. In comparison, the maps of the MultiNet GRAPPA and the fully sampled data look very similar. No visible aliasing artifacts are present in the MultiNet GRAPPA lipid contamination maps.
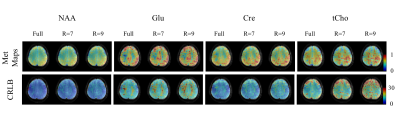
Figure 4 shows the metabolite maps for the fully sampled, along with the highly accelerated R=7 and R=9 MultiNet PyGRAPPA reconstructions. Compared to the fully sampled data, the accelerated maps show good consistency albeit slightly more noisy. The grey/white matter contrast and the anatomical structure can still be seen. Despite the high in-plane acceleration factors no visible residual aliasing artifact can be discerned.
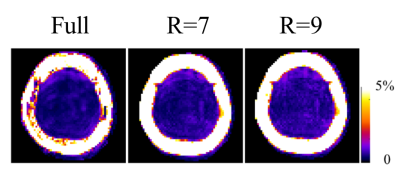
Figure 5 shows the lipid contamination maps for the R=7 and R=9 MultiNet PyGRAPPA reconstructions. Despite the high in-plane acceleration factors, there are no visible aliasing artifacts and lipid rings in these maps.
Conclusion
In this work, we present a novel acquisition and reconstruction scheme (MultiNet PyGRAPPA) for highly accelerated metabolite mapping using non-lipid suppressed 1H FID MRSI. The proposed method combines a variable density sampling scheme with multiple neural networks for reconstruction. The machine learning based reconstruction method proves superior to conventional GRAPPA and results in less noise and residual aliasing artifacts. The proposed method was applied at 9.4T for high resolution (matrix size = 64x64) metabolite mapping of the human brain acquired in 2.8 or 2 minutes, for R=7 and 9, respectively.Acknowledgements
No acknowledgement found.References
[1] Strasser et al, MRM 2016.
[2] Hangel et al, MRM 2015.
[3] Avdievitch et al, NMR in Biomed 2017.
[4] Nassirpour et al, NeuroImage 2016.
[5] Henning et al, NMR in Biomed 2009.
[6] Bogner et al, NMR in Biomed 2012.
[7] Blaimer et al, MRM 2006.
Figures