3370
Compressed Sensing MRI Reconstruction using Generative Adversarial Networks with Cyclic Loss.1School of Electrical Computer Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan, Republic of Korea
Synopsis
Compressed Sensing MRI (CS-MRI) has provided theoretical foundations upon which the time-consuming MRI acquisition process can be accelerated. However, it primarily relies on iterative numerical solvers which still hinders their adaptation in time-critical applications. In addition, recent advances in deep neural networks have shown their potential in computer vision and image processing, but their adaptation to MRI reconstruction is still at an early stage. Therefore, we propose a novel compressed sensing MRI reconstruction algorithm based on a deep generative adversarial neural network with cyclic data consistency constraint. The proposed method is fast and outperforms the state-of-the-art CS-MRI methods by a large margin in running times and image quality, which is demonstrated via evaluation using several open-source MRI databases.
Introduction
The primary motivation for the proposed work stems from the following observations: recent advances in deep learning can “shift” the time-consuming computing process into the training (pre-processing) phase and save prediction time by performing only one-pass deployment instead of using iterative methods. The proposed model is a variant of fully-residual convolutional autoencoder and generative adversarial networks (GANs)1, specifically designed for CS-MRI formulation; it employs cyclic data consistency losses2 for faithful interpolation of the given under-sampled k-space data. In addition, our solution leverages a chained network to further enhance the reconstruction quality. The proposed method is fast and accurate - the reconstruction process is extremely rapid, because it is one-way deployment on a feed-forward network, and the image quality is superior even for extremely low sampling rate (as low as 10%) due to the data-driven nature of the method.Method
Figure 1 represents an overview of the proposed method: Our generator G consists of two-fold chained networks that try to reconstruct the MRI image directly from under-sampled k-space(zero-filled reconstruction). The generated result is favorable to the fully-sampled data taken from an extensive database and put through the same under-sampling process. In contrast, the discriminator D attempts to differentiate between the real MRI instances from the database and the fake results output generated by G.
Figure 2 is a schematic depiction of our adversarial process: G tries to generate images s_ = G(m) that look similar to the images s that have been reconstructed from full k-space data, while D aims to distinguish between s_ and s. Once the training converges, G can produce the result s_ that is close to s, and D is unable to differentiate between them, which results in bringing the probability for both real and fake labels to 50%.The cyclic data consistency loss, which is a combination of under-sampled frequency loss and the fully reconstructed image loss helps to strengthen the bridge connection between zero-filling and fully-sampled reconstruction of one instance.
Results
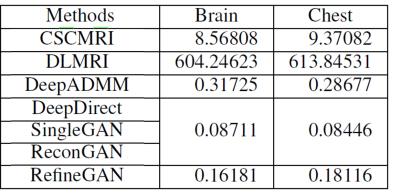
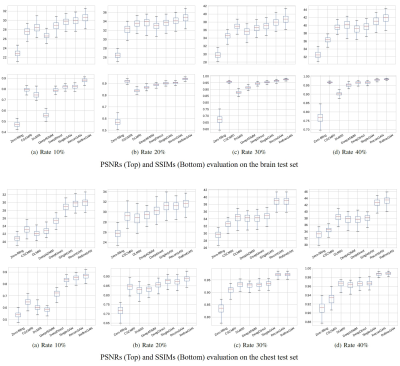
We used two sets of MR images from the brain dataset and from the chest dataset to assess the performance of our method by comparing our results with state-of-the-art CS-MRI methods (e.g., convolutional sparse coding-based (CSCMRI3,4), patch-based dictionary (DLMRI5,6,7), deep learning-based (DeepADMM8; DeepDirect9,10), and GAN-based (SingleGAN11,12). The resolution of each image is 256x256. From each database, we randomly selected 100 images for training the network and another 100 images for testing the result. We conducted the experiments for various sampling rates (i.e., 10%, 20%, 30%, and 40% of the original k-space data) under the radial sampling strategy.
Figure 3 summarizes the running times of our method and other state-of-the-art learning-based CS-MRI methods. The running times of DeepDirect, SingleGAN and our ReconGAN are, all similarly, about 0.08 second because they share the same network architecture (i.e., single fold generator G). The running time of RefineGAN is about twice as long because two identical generators are serially chained in a single architecture, but it is still around 0.16 second. As shown in this experiment, we observed that deep learning based approaches are well-suited for CS-MRI in a time-critical clinical setup due to their extremely low running times.
Discussion
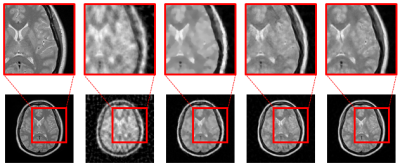
By qualitatively comparing the reconstruction results (Figure 4 and 5), we found that deep learning-based methods generate more natural images than dictionary-based methods. For example, CSCMRI and DLMRI produce cartoon-like piecewise linear images with sharp edges, which is mostly due to sparsity enforcement. In comparison, our method generates results that are much closer to full reconstructions while edges are still preserved; in addition, noise is significantly reduced. Note also that, unlike other CS-MRI methods, our method can generate acceptable results even at extremely low sampling rates.Conclusion
The proposed architecture is specifically designed to have a deeper generator network G and is trained adversarially with the discriminator D with cyclic data consistency loss to promote better interpolation of the given undersampled k-space data for accurate end-to-end MR image reconstruction. We demonstrated that our method outperforms the state-of-the-art CS-MRI methods in terms of running time and image quality. In the future, we plan to conduct an in-depth analysis of the proposed method to better understand the architecture, as well as constructing incredibly deep multi-fold chains with the hope of further improving reconstruction accuracy based on its target-driven characteristic. Extending our method to handle dynamic MRI is an immediate next research direction. Developing a distributed version of the proposed method for parallel training and deployment on a cluster system is another research direction we wish to explore.
Acknowledgements
This work is partially supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1A09000841), the Bio & Medical Technology Development Program of the NRF funded by the Ministry of Science & ICT (NRF-2015M3A9A7029725), and the 2017 Research Fund (1.170017.01) of UNIST (Ulsan National Institute of Science and Technology).References
1. I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, and Y. Bengio, “Generative Adversarial Nets,” in Proceeding of NIPS, 2014.
2. J.-Y. Zhu, T. Park, P. Isola, and A. A. Efros, “Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks,” in Proceeding of ICCV, 2017.
3. T. M. Quan and W.-K. Jeong, “Compressed sensing dynamic MRI reconstruction using GPU-accelerated 3D convolutional sparse coding.” in Proceeding of MICCAI, 2016.
4 T. M. Quan and W.-K. Jeong, “Compressed sensing reconstruction of dynamic contrast-enhanced MRI using GPU-accelerated convolutional sparse coding,” in Proceeding of IEEE ISBI, 2016.
5. S. Ravishankar and Y. Bresler, “MR image reconstruction from highly undersampled k-space data by dictionary learning,” IEEE Transactions on Medical Imaging, vol. 30, no. 5, pp. 1028–1041, 2011.
6. J. Caballero, D. Rueckert, and J. V. Hajnal, “Dictionary learning and time sparsity in dynamic MRI,” in Proceeding of MICCAI, 2012.
7. J. Caballero, A. N. Price, D. Rueckert, and J. V. Hajnal, “Dictionary learning and time sparsity for dynamic MR data reconstruction,” IEEE Transactions on Medical Imaging, vol. 33, no. 4, pp. 979–994, 2014.
8. J. Sun, H. Li, Z. Xu et al., “Deep ADMM-net for compressive sensing MRI,” in Proceeding of NIPS, 2016.
9. S. Wang, Z. Su, L. Ying, X. Peng, S. Zhu, F. Liang, D. Feng, and D. Liang, “Accelerating magnetic resonance imaging via deep learning,” in Proceeding of IEEE ISBI, 2016
10. D. Lee, J. Yoo, and J. C. Ye, “Deep residual learning for compressed sensing MRI,” in Proceeding of IEEE ISBI, 2017.
11. M. Mardani, E. Gong, J. Y. Cheng, S. Vasanawala, G. Zaharchuk, M. Alley, N. Thakur, S. Han, W. Dally, J. M. Pauly et al., “Deep Generative Adversarial Networks for Compressed Sensing AutomatesMRI,” arXiv preprint arXiv:1706.00051, 2017.
12. S. Yu, H. Dong, G. Yang, G. Slabaugh, P. L. Dragotti, X. Ye, F. Liu, S. Arridge, J. Keegan, D. Firmin et al., “Deep De-Aliasing for FastCompressive Sensing MRI,” arXiv preprint arXiv:1705.07137, 2017.
Figures
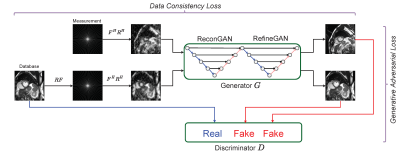
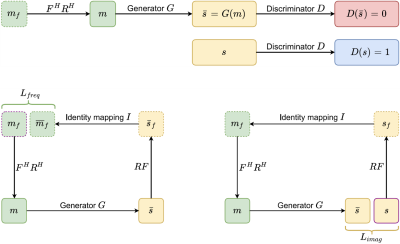
(Top) Two learning processes are trained adversarially to achieve better reconstruction from generator G and to fool the ability of recognizing the real or fake MR image from discriminator D.
(Bottom) The cyclic data consistency loss, which is a combination of under-sampled frequency loss and the fully reconstructed image loss.