3366
MR Image Generation with Deep Learning Incorporating Anatomical Prior Knowledge1Department of Bio and Brain Engineering, Korea Advanced Institute of Science & Technology (KAIST), Daejeon, Republic of Korea
Synopsis
We proposed a new convolutional neural network (CNN) to generate high resolution (HR) MR images from highly down-sampled MR images, incorporating HR images in another contrast. Anatomical information from another HR images and adversarial loss functions allowed the proposed model to restore details and edges clearly from the down-sampled images, proved in normal and brain tumor regions. Pre-training with a public database improved performance in real human applications. The proposed methods outperformed several CS algorithms in both pseudo-k-spaces from public data and real k-spaces from human brain data. CNNs can be a good alternative for accelerating routine MRI scanning.
Introduction
Since high frequency contents are not significantly different irrespective of MRI pulse sequence, anatomical information from one full-sampled high-resolution (HR) image can improve another down-sampled low-resolution (LR) image, reducing the total scan time. In this study, we propose a new convolutional neural network (CNN) to generate HR MR images from highly down-sampled MR images, by incorporating another HR MR images acquired with a different pulse sequence. In addition, adversarial loss is proposed to improve perceptual image quality and pre-training with public data before fine-tuning with real k-space data is proposed to improve the performance further.Methods
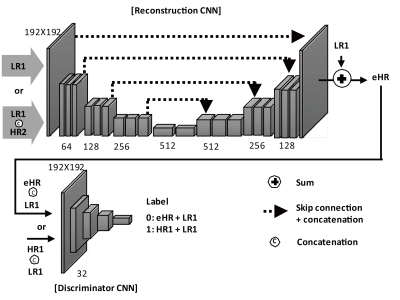
The proposed models consisted of two CNNs: (1) a reconstruction CNN for generating the HR image from the down-sampled images using another HR image acquired with a different pulse sequence and (2) a discriminator CNN for differentiating between the generated HR image and the full-sampled HR image called “ground truth” (Figure 1a). The two CNNs, adversarial to each other in nature, were combined in a specially-designed loss function for better perceptual image quality. The reconstruction CNN was based on a combination of Unet [1] and residual training scheme [2].
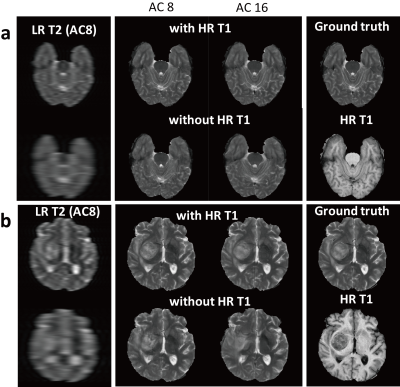
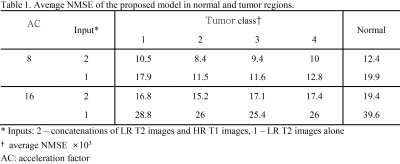
CNN were evaluated with pseudo k-spaces from BRATS database [3]. The central 48 and 16 lines along the first dimension were chosen to produce 8- and 16-fold down-sampled T2 images, respectively. The down-sampled T2 images and HR T1 images were used as inputs for the proposed model. The performance of the proposed model was evaluated with normalized mean square error (NMSE) in normal and tumor regions. Region-of-interests (ROIs) of brain tumor that were provided by BRATS organizer were used to quantify the error in tumor regions.
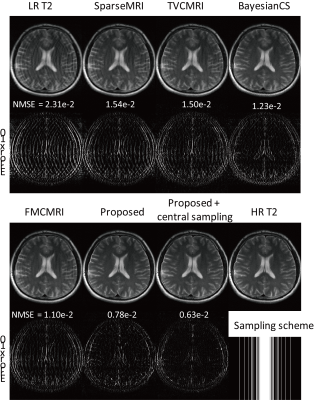
CNNs were applied to brain MRI datasets acquired from ten subjects with Cartesian coordinate on a 3T MRI scanner (MAGNETOM Trio, Siemens Healthcare, Erlangen, Germany). Axial double echo T2- and proton density (PD)-weighted images were obtained with the following parameters: repetition time, 3700 ms; echo time, 37/103 ms; resolution, 0.7 × 0.7 mm2 and thickness, 5 mm. Also, axial spin-echo T1-weighted images were acquired with the same parameters except for repetition time of 500 ms and echo time of 9.8 ms. The models pre-trained with BRATS data were fine-tuned using the down-sampled T2/PD images and HR T1 images from three subjects. The performance of the fine-tuned models was compared to those of several compressive sensing (CS) algorithms using datasets from the remaining seven subjects.
Results
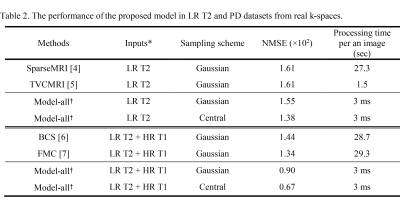
CNNs trained with HR T1 images showed better perceptual image quality than those without HR T1 images in normal and tumor regions, irrespective of accelerating factor (Figure 2). Also, CNNs with HR T1 images showed lower NMSE values than those without HR T1 images (Table 1). Although tumor was only part of the whole data, CNNs with HR T1 images could restore details of tumor well, probably with the help of high frequency contents of the HR T1 images. The results from the seven k-space datasets were consistent with those from the BRATS datasets (Figure 3). The proposed model significantly decreased the error, especially upon combination with HR T1 images. As compared to CNNs without pre-training with BRATS data (NMSE = 1.1 in PD and 2.2 in T2), CNNs with combination of pre-training with BRATS datasets and fine-tuning with three k-space datasets improved NMSE (NMSE = 0.8 in PD and 1.6 in T2). CNNs provided lower NMSE values (Table 2) and perceptually clearer MR images than the CS algorithms (Figure 4). The processing time of the proposed method was also faster than the CS algorithms (Table 2). In the proposed model, the central sampling scheme resulted in lower NMSE values than the Gaussian sampling scheme.Discussion and Conclusion
We proposed a deep learning approach to generate high-resolution MR images from highly down-sampled LR MR images in normal and brain tumor regions, by incorporating (i) anatomical information from another HR images acquired with a different MR sequence and (ii) the adversarial loss function. Incorporation of another HR images and the adversarial loss function allowed the proposed model to restore details and edges clearly from the low-resolution images, proved in normal and brain tumor regions. Also, transfer learning using public datasets and fine-tuning with datasets from a small number of subjects improved the proposed methods for the real application, with demonstrated superiority over the CS algorithms. The proposed method can be a good strategy for accelerating MR imaging.Acknowledgements
This work was supported by the National Research Foundation of Korea (2017R1A2B2006526) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare of South Korea (HI16C1111).References
1. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv preprint. 2015;arXiv:1505.04597.
2. Lee D, Yoo J, Ye JC. Deep artifact learning for compressed sensing and parallel MRI. arXiv preprint. 2017;arXiv:1703.01120.
3. Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE transactions on medical imaging. 2015;34(10):1993-2024.
4. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnet Reson Med. 2007;58(6):1182-95.
5. Ma S, Yin W, Zhang Y, Chakraborty A. An efficient algorithm for compressed MR imaging using total variation and wavelets CVPR. 2008. 1-8 p.
6. Bilgic B, Goyal VK, Adalsteinsson E. Multi-contrast reconstruction with Bayesian compressed sensing. Magn Reson Med. 2011;66(6):1601-15.
7. Huang J, Chen C, Axel L. Fast multi-contrast MRI reconstruction. Magn Reson Imaging. 2014;32(10):1344-52.
Figures