3363
Densely Connected Iterative Network for Sparse MRI Reconstruction1GE Global Research, Herzliya, Israel, 2GE Global Research, Niskayuna, NY, United States, 3GE Healthcare, Waukesha, WI, United States, 4GE Healthcare, Menlo Park, CA, United States
Synopsis
We propose a densely connected deep convolutional network for reconstruction of highly undersampled MR images. Eight-channel 2D brain data with fourfold undersampling were used as inputs, and the corresponding fully-sampled reconstructed images as references for training. The algorithm produced notably higher-quality images than state-of-the-art parallel imaging and compressed sensing methods, both in terms of reconstruction error and perceptual quality. The dense architecture was found to significantly outperform a similar network without dense connections.
Purpose
Parallel imaging and compressed sensing have been used to reduce MRI scan times through undersampling of k space, exploiting the different spatial sensitivities of coil arrays and/or sparsity in some transform domain. Recently, deep-learning techniques have been brought to bear on this problem [e.g., 1-3], producing improved image quality for similar undersampling factors. We propose a new architecture, called DCI-Net (Densely Connected Iterative Network), based on an iterative convolutional network, to bring further improvements in image quality for sparse MR image reconstruction. The key new developments are the use of 1) dense connections [4] across all iterations, which strengthens feature propagation, making the network more robust, and 2) a relatively deep architecture of over 100 convolutional layers, bringing increased capacity.Methods
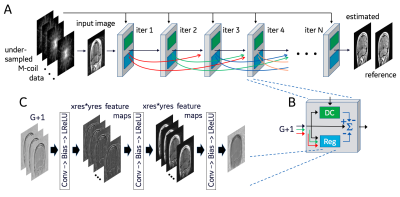
The DCI-Net (Fig. 1), receives M coils of undersampled k-space data at its input. The network is an unrolled compressed-sensing (CS) iterative reconstruction with N = 40 iterations, each of which includes a data-consistency unit and a regularization unit (Fig. 1B). Skip-layer connections between the output of each iteration and the following G iterations - where typically G = 16 - are represented as curved lines in Fig. 1A. This results in an input to each block composed of skip and direct connections concatenated to form a G+1 channel complex image. (The number of skip connections is ramped down in the final iterations until there is only one channel at the output of the network.) Each data-consistency unit shades the input image with each coil sensitivity map, transforms the resulting images to k-space, imposes the sampling mask, calculates the difference relative to acquired k-space and returns them to the image domain, multiplied by a learned weight. Each regularization unit (Fig. 1C) has three sequences consisting of 5x5 convolution, bias, and leaky-ReLU layers. The output of the final iteration (Fig. 1A) is converted to a magnitude image and compared to the fully sampled reference image to generate a loss function, using either mean square error (MSE) or structural similarity index (SSIM).
Fully sampled multi-slice brain datasets (256x256 T1-FLAIR and T2-FLAIR, in axial, coronal, and sagittal orientations), along with separately acquired sensitivity maps, were acquired after informed consent from 12 healthy volunteers at 3T using an 8-channel receive array. The data were retrospectively down-sampled using 11 central lines of k-space and a fixed pseudo-random sampling pattern outside the central region, resulting in a net undersampling factor of 3.6. Complex coil-combined zero-filled reconstructions were used as input to the network. The undersampled k-space data were also input directly into each iterative block of the network (Fig. 1A) for use in the data-consistency units. The fully sampled k-space data were reconstructed to form reference images for comparison with the network output. In total, 1130 slices were collected, of which 948 were used to train the networks, 89 for validation and 93 for testing. The same undersampled data were also reconstructed using compressed sensing (with both total variation and wavelets). Parallel imaging (ARC) reconstructions were performed as well, but with regular undersampling in outer k space.
Results
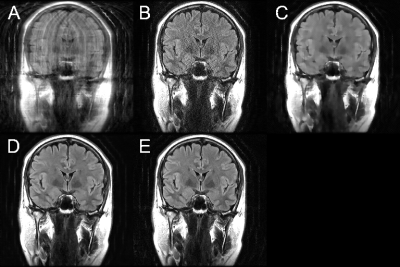
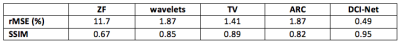
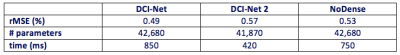
Figure 2 shows an example of one of the test images, comparing the output of the trained DCI-Net with the zero-filled image, the fully sampled image, and two alternative sparse reconstructions. Table 1 gives a comparison of mean image quality for the various methods, from the test dataset. It can be seen that the DCI-Net produces significantly more accurate reconstructions than the other methods. Table 2 shows mean relative MSE (rMSE), number of learned parameters, and inference runtime for the same test dataset, comparing the DCI-Net with two other configurations. Employing dense connections has significantly improved accuracy, and the use of the deep network produced 16% lower mean rMSE than a shallower network, while using a similar number of learned parameters.Discussion
DenseNets [4] differ from a prior skip-layer method [5] in that they employ skip connections to connect with all neighboring layers, and combine features through concatenation, not summing. Dense networks are useful for alleviating the vanishing-gradient problem, and for strengthening feature propagation and reuse, allowing much deeper networks than would otherwise be possible. We have found that adding dense connections and increasing the depth of the network significantly improves performance of our sparse MRI reconstruction.Conclusion
This work presents a novel architecture for sparse MRI reconstruction based on iterative deep convolutional networks, that densely connects all iterations, producing significantly higher image quality. This should enable sparser sampling and thus higher clinical imaging speeds.Acknowledgements
No acknowledgement found.References
1. J Schlemper, et al. A deep cascade of convolutional neural networks for MR image reconstruction. arXiv:1703.00555v1, 1 March 2017.
2. K Hammernik, et al. Learning a variational network for reconstruction of accelerated MRI data. arXiv:1704.00447v1, 3 Apr 2017.
3. S Diamond, et al. Unrolled optimization with deep priors. arXiv:1705.08041v1, 22 May 2017.
4. G Huang, et al. Densely connected convolutional networks. arXiv:1608.06993v4, 27 Aug 2017 (Proceedings of CVPR 2017).
5. K. He, X. Zhang, S. Ren, and J. Sun. Deep residual learning for image recognition. arXiv:1512.03385v1, 10 Dec 2016 (Proceedings of CVPR 2016).
Figures