3356
Highly Accelerated 3D MR Angiography Using Multi-Channel Blind Deconvolution1Biomedical Engineering, State University of New York at Buffalo, Buffalo, NY, United States, 2United-Imaging Healthcare America, Houston, TX, United States, 3Electrical Engineering, State University of New York at Buffalo, Buffalo, NY, United States, 4The MRI Institute for Biomedical Research, Detroit, MI, United States, 5Department of Radiology, Wayne State University, Detroit, MI, United States
Synopsis
In many clinical applications, the three dimensional (3D) MRA plays an important role because that the 3D MRA can provide plenty of details for more compact anatomic regions with various flow directions. However, the speed limitation of the 3D MRA reconstruction is still an unignorable problem due to the size of the dataset, especially when the dataset has multi channels. With our proposed method, the Multi-Channel Blind Deconvolution (MalBEC), the experiment demonstrate that this method can provide high quality reconstruction image with high acceleration factors using much less time.
Purpose
Understanding the cerebral vascular system is very important in many clinical applications. Three dimensional (3D) MRA using bright-blood strategies such as time-of-flight (TOF) have been widely used to obtain high blood signal with high spatial resolution and high signal-to-noise ratio while reducing tissue signal as much as possible. However, the imaging time is much longer than 2D MRA techniques. To accelerate 3D MRA acquisition, compressed sensing [1] has been investigated and shown to be able to achieve an acceleration factor of 5 without compromising the image quality [2]. Recently we have proposed the Multi-Channel Blind Deconvolution (MalBEC) method [3] to reconstruct 3D images from highly undersampled data. The objective of this work is to study how much MalBEC can accelerate 3D MRA acquisitions.Methods
The MalBEC formulates the image reconstruction problem from multi-channel undersampled data as to recover the unknown k-space data in all channels using blind deconvolution. An important feature of MalBEC is that it implicitly utilizes the coil sensitivity information but without the need to estimate it a priori. Different from the existing methods where either sparsity constraints are applied in image domain (e.g., [4-7]) or rank deficiency is applied in k-space (e.g., [8]) to estimate the coil sensitivity information indirectly, MalBEC assume the coil sensitivities are smooth such that they have limited support in k-space. The assumption is generally valid for most phased array coils. In addition, MalBEC is highly efficient in computation and is requires much shorter CPU time than the existing methods for 3D reconstruction. Specifically, we solve the desired k-space data s and channel responses hc alternatively:
s step:$$$s=arg\min _{s}\sum_c||y_{c}-Ω(s\circledast h_{c})||^{2}$$$ (1)
h step:$$$h_{c} = arg\min_{h_{c}}||y_{c}-Ω(s\circledast h_{c}||^{2}$$$ (2)
where yc is acquired data and Ω as the undersampling operation. Those two steps are performed alternately and iteratively. After convergence, Fourier transform of the k-space data s provides the desired image.
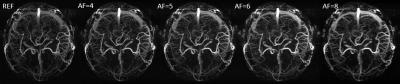
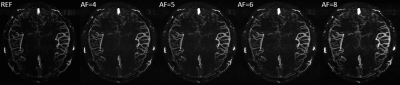
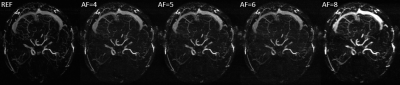
We applied MalBEC on 3D MRA datasets. A healthy volunteer was recruited for the study and written consents approved by the local Institutional Review Board were obtained from the volunteer prior to the scans. All scans were performed on a Siemens 3T Verio system (Siemens Healthcare, Erlangen, Germany) with a product 32-channel head coil. An interleaved three-echo GRE sequence [9,10] was used for acquisition where three images were acquired within a single scan. The scanning parameters of the interleaved three-echo GRE sequence were: TE1/TE2(flow-rephased)/TE3(flow-dephased)/TR = 2.5/13/13/20 msec, flip angle (FA)= 12, bandwidth = 260 Hz/pixel, voxel size = 0.67 × 0.67 × 1.2 mm3, matrix size = 384 x 288 x 104. The two flow-rephased and flow-dephased images were subtracted to generate an MRAV. To evaluate the performance of MalBEC under different acceleration factors of 4, 5, 6, and 8, we retrospectively undersample the 3D data with 2D pseudo-random mask [11] (in phase encoding and partition encoding directions).
Results
The maximum intensity projections (MIP) of the reconstructed 3D MRAV results with difference acceleration factors are shown in Figure 1, 2 and 3. It can be seen that MalBEC can recover all the fine details at even a high acceleration factor of 8. The average reconstruction time is 20 minutes for each echo/image.Conclusion
The MalBEC method can reach a high quality images with high acceleration factors. And this method is also highly efficient in terms of computational complexity which can make 3D online reconstruction possible.Acknowledgements
This work is supported in part by the NSF CCF-1514403, NIH R21EB020861.References
1. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58:1182–1195.
2. Fushimi Y, Fujimoto K, Okada T, Yamamoto A, Tanaka T, Kikuchi T, Miyamoto S, Togashi K. Compressed Sensing 3-Dimensional Time-of-Flight Magnetic Resonance Angiography for Cerebral Aneurysms: Optimization and Evaluation. Invest Radiol. 2016 Apr;51(4):228-35.
3. Lyu J, Nakarmi U, Zhou Y, Zhang C, Ying L. Calibration-free Parallel Imaging Using Randomly Undersampled Multichannel Blind Deconvolution (MALBEC). ISMRM, 3232, 2016
4. Liu B, Zou YM, Ying L, editors. SparseSENSE: application of compressed sensing in parallel MRI. Information Technology and Applications in Biomedicine, 2008 ITAB 2008 International Conference on; 2008: IEEE.
5. Liang D, Liu B, Wang J, Ying L. Accelerating SENSE using compressed sensing. Magnetic Resonance in Medicine. 2009;62(6):1574-84.
6. She H, Chen RR, Liang D, DiBella EV, Ying L. Sparse BLIP: BLind Iterative Parallel imaging reconstruction using compressed sensing. Magnetic Resonance in Medicine. 2014;71(2):645-60.
7. Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010 Sep;64(3):767-76. doi: 10.1002/mrm.22463.
8. Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med. 2010;64(2):457-71.
9. Ye Y, Hu J, Wu D, Haacke M. Noncontrast-enhanced magnetic resonance angiography and venography imaging with enhanced angiography. J Magn Reson Imaging 2013; 38: 1539–1548.
10. Chen Y, Liu S, Kang Y, and Haacke EM. An interleaved sequence for simultaneous MRA, SWI and QSM. ISMRM 2017, p1215.
11. Kim W, et al. Conflict-cost based random sampling design for parallel MRI with low rank constraints. SPIE Sensing Technology+ Applications. International Society for Optics and Photonics, 2015.
Figures