3270
Construction of an MRI-Based Connectome for the Marmoset Brain: Methods and Initial ResultsCecil Chern-Chyi Yen1, Cirong Liu1, and Afonso C. Silva1
1National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States
Synopsis
The common marmoset, a small New World primate, is an excellent translational animal model due to the similarity between its brain network and humans. However, marmoset’s brain networks are not fully explored because of lacking a large group of resting-state functional MRI (
Introduction
The human connectome project (HCP) is the most successful multi-center collaboration efforts aimed at mapping neural connections of the human brain1. The success of the HCP may be attributed to having standardized MRI protocols that are optimized for instruments of various centers. A pilot version of the HCP-style protocol has been developed for 7T2. Ultra-high field MRI provides unprecedented spatial resolution and sensitivity, which can be combined to facilitate detection of finer and hidden brain connections. These brain networks obtained from healthy subjects build a solid baseline for comparative studies of patients with either acquired or congenital brain disorders. However, to investigate the etiology and treatments of such disorders, an animal model whose brain networks and gnome share a high degree of similarity to humans is desired. The common marmoset, a small New World primate, has shown to pose these two properties. In addition, germline transgenesis in marmosets opens the opportunity to create a non-human primate model of many congenital brain disorders3. Despite the enormous potential of the marmoset model, a population-averaged baseline of marmoset’s brain connections is still lacking. Our group is one of the pioneers in resting-state functional MRI (rfMRI) of awake marmosets4. The use of awake marmosets in functional imaging studies eliminate the confounds of general anesthesia, that hampers the detectability and interpretation of brain networks and increases data variability across multi-centers. Therefore, the aim of this study is to create a standardized MRI protocol for constructing the marmoset connectome, that will hopefully promote interinstitutional collaborations.Methods
All procedures were approved by the ACUC of the NINDS. Seven adult male marmosets (age: 2-7 years old) were acclimated to the body and head restraint inside a horizontal 7T/30cm MRI spectrometer (Bruker., Billerica, USA). Marmosets were laid in the sphinx position in the cradle, and their heads were comfortably immobilized by individualized 3D-printed helmet and chin-piece5. Under sphinx position, marmoset’s body would be lower than its head and would not fit into 9cm volume transmission coil normally came with 12cm gradient coil. Thus, a 15cm gradient coil (RRI., Billerica, USA) was used with an 11cm custom-built birdcage coil. A custom-built 10-element phased array RF coil was placed on top of the helmet. FLASH and RARE sequence were used to align three-orthogonal planes to the mid-sagittal plane and the line connecting anterior and posterior commissure. Anatomical MRI data were acquired using a RARE sequence from 38 coronal slices with 0.5 mm isotropic resolution. A pair of spin-echo EPI with opposite phase-encoding gradient were acquired for correction of the susceptibility-induced off-resonance field. rfMRI data were acquired using a single-shot gradient-echo EPI sequence. Four to six rfMRI scans with 512 repetitions were performed in each session. MRI protocols and parameters can be found in figure 1. In figure 2, pre- and post-processing of the data were done using AFNI, FSL and ANTS, which can be incorporated into an unattended batch script and register to NIH’s marmoset brain atlas6.Results
In figure 2, study-specific anatomical MRI images show excellent image quality and tissue contrast. The slice thickness and number were deliberately kept the same as the rfMRI for easier importing geometry. The co-planar rfMRI images achieve good SNR (>20 for all images) and temporal stability. EPI distortion due to the susceptibility-induced off-resonance field was minimized using a pair of blip -up and -down EPI and FSL’s topup. Three major hubs (dlPFC, PCC, and PCRSC) of default mode network are observed in group averaged template (figure 3) and all seven marmosets (figure 4).Discussion
Our MRI protocol and processing pipeline of marmoset connectome follow the well-considered HCP-style MRI protocol of 7T and its processing pipeline. Due to the difference in marmoset vs. human brain and pre-clinical vs. clinical MRI, exceptions and limitations are inevitable. First, the spatial resolution was determined to be 0.5mm isotropic, which was the best resolution achievable with acceptable SNR. Second, we chose the longest axis (anterior-posterior) as our frequency-encoding direction and left-right axis as our phase-encoding direction for parallel imaging. The slice-selection direction is along the shorter dorsal-ventral axis and hence limits multiband capability. Third, our TR is set to 2s due to the limitation of the gradient duty cycle without multiband acceleration. In conclusion, we developed an MRI protocol for imaging awake marmosets following the methodology of the HCP. Our MRI protocol and process pipeline can be easily adopted across centers currently interested in imaging the marmoset brain. We believe our standardized MRI protocol will encourage the development of global collaborations, and lead to a further utilization of the marmoset model in translational neuroscience studies.Acknowledgements
We thank Lisa Zhang and Madeline Marcelle for animal preparation and training. We also thank Dr. Hanbing Lu for providing reverseEPI sequence. This research was supported by the Intramural Research Program of the NINDS.References
- Van Essen, D. C. et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62-79, doi:10.1016/j.neuroimage.2013.05.041 (2013).
- HCP Protocols, <http://protocols.humanconnectome.org/> (2014).
- Park, J. E. et al. Generation of transgenic marmosets expressing genetically encoded calcium indicators. Sci Rep 6, 34931, doi:10.1038/srep34931 (2016).
- Belcher, A. M. et al. Large-scale brain networks in the awake, truly resting marmoset monkey. J Neurosci 33, 16796-16804, doi:10.1523/JNEUROSCI.3146-13.2013 (2013).
- Papoti, D., Yen, C. C., Mackel, J. B., Merkle, H. & Silva, A. C. An embedded four-channel receive-only RF coil array for fMRI experiments of the somatosensory pathway in conscious awake marmosets. NMR Biomed 26, 1395-1402, doi:10.1002/nbm.2965 (2013).
- Liu, C. et al. A digital 3D atlas of the marmoset brain based on multi-modal MRI. Under revision (2017).
Figures

Figure 1. MRI protocol of the marmoset connectome. In MRI protocol, orange is FLASH sequence; green is RARE sequence; gray is filed map; blue is EPI sequence. Multiple localizers and scouts might be required for positioning and aligning. Anatomical MRI: TEeff/TR= 54/6000ms; in-plane resolution=0.25mm, slice thickness=0.5mm; matrix=144×112, Bandwidth=25KHz, GRAPPA=2 with 32 reference lines, 8 averages. Functional MRI: TE/ TR= 22.2/ 2000ms; spatial resolution=0.5mm isotropic; matrix=72×56, Bandwidth=134KHz, GRAPPA=2, Double sampling.

Figure 2. Processing pipeline of the marmoset connectome. Orange is AFNI; green is FSL; blue is ANTS; gray is depending on users. To simplify the flowchart, brain masks generated by multi-atlas fusion brain extraction are only applied during construction of study-specific template.
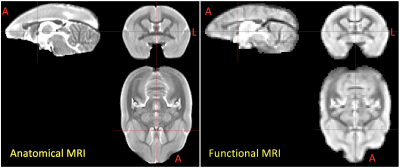
Figure 3. Population-averaged anatomical
(RARE) and functional (EPI) MRI data of awake marmosets. Good quality of data
is observed demonstrating the optimization of the MRI protocol. Red cross-hairs
are the center at the top of the anterior commissure. Marmoset’s brain was
aligned perfectly by the line between the top of AC and the bottom of PC and
mid-sagittal plane. A: anterior, L: left
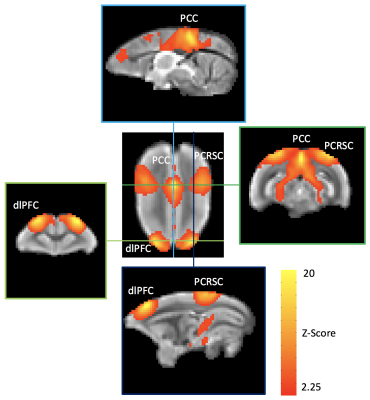
Figure
4. Default mode network of awake marmosets generated by group
ICA. Robust default mode network is observed in group ICA. Horizontal and
vertical lines show interceptions of two coronal and sagittal slices with
distinct correlation pattern. Marmoset’s default mode network is very similar
to those reported in human, except slight different in PFC area. dlPFC: dorsal-lateral prefrontal
cortex,
PCC: posterior parietal cortex,
PCRSC: posterior cingulate and retrosplenial cortex
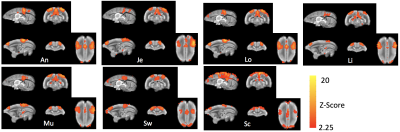
Figure 5. Default mode network of seven awake marmosets generated by ICA. The default mode network is consistently observed in all marmosets.