3267
Investigation of the default mode network in awake marmosets1National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 2National Institute of Mental Health, National Institutes of Health, Bethesda, MD, United States
Synopsis
We investigated the default mode network (DMN) in awake marmosets, by combining both resting-state and task-based fMRI, and by performing diffusion tractography and neuronal tract tracing. We found that dorsolateral (dlPFC) but NOT medial prefrontal cortex (mPFC) is a major region of the marmoset DMN. The dlPFC has direct anatomical connections and strong functional connectivity with the posterior DMN and also exists in macaques and humans, suggesting that it is more evolutionary conservative than the mPFC. However, its functions as a default mode are poorly understood and worth investigation.
INTRODUCTION
The default mode network (DMN), a large-scale brain network that deactivates during active, has attracted considerable interest for understanding the significance of the brain’s intrinsic activity. To clarify both anatomical definition and functional significance of the cortical areas involved in the DMN, it is imperative to investigate the DMN in non-human primate models that allow more direct methods to probe the DMN. In this study, we systematically investigated both anatomical and functional connectivity of the DMN in marmosets, a valuable non-human primate model with great potential in neuroscience. We hope our comparative analyses will provide new insights into the significance and function of the DMN.METHODS
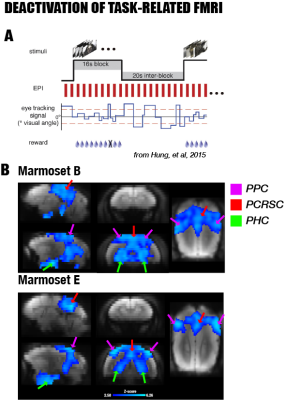
This study recruited seven adult male marmosets that underwent resting-state fMRI (0.5mm isotropic, TR=2s, 4-6 runs with 512 time points per run using a newly-designed 10-channel surface coil at 7T) and two other marmosets that underwent a visual task fMRI 1 (0.5mmx0.5mmx1mm, TR=2s; a block design with different visual stimuli using an 8-channel coil at 7T). After data preprocessing, the resting-state fMRI was used to locate the correlated cortical areas of the DMN by group independent component analysis (ICA), while the task-based fMRI was used to examine deactivation (defined as regions showing negative response patterns) during the visual task. By registering fMRI maps to the MRI templates of the NIH Marmoset Brain Atlas 2, we determined the corresponding locations of these cortical components and examined their anatomical connections by querying a retrograde neuronal tracing database of the marmoset brain 3-4. As the retrograde neuronal tracing did not reveal fiber pathways, we further performed probabilistic tractography using ultra-high resolution ex-vivo diffusion MRI (150µm isotropic resolution at 7T) and reconstructed the white matter tracts underlying the DMN of the marmoset brain.RESULTS
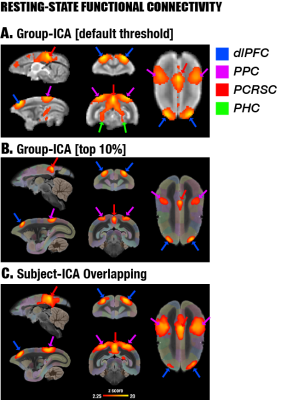
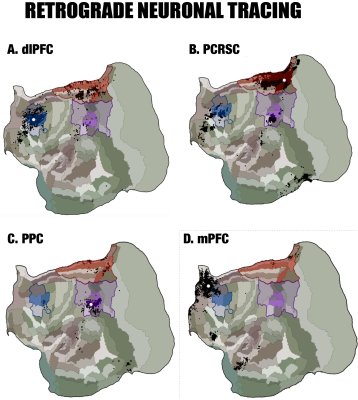
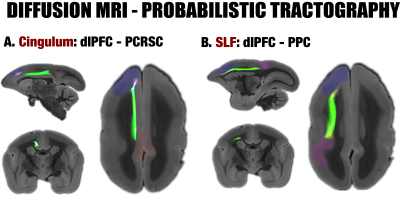
By performing resting-state fMRI in awake marmosets, we found that the dorsolateral prefrontal cortex (dlPFC; including areas 8aD and 6DR), but NOT the medial prefrontal cortex (mPFC), was a major region of the marmoset DMN (Fig.1). The other two major regions of DMN were the posterior cingulate and retrosplenial region (PCRSC; including areas PGM, A30, A29d) and the posterior parietal cortex (PPC; including the intraparietal area). Hippocampal and para-hippocampal regions (PHC) were also involved in the DMN but not as prominent as the three main hubs above. A small area near mPFC (A32) was also observed as a weak component, but it was more likely to be a false-positive due to vascular artifacts in nearby areas, as its peak was located outside the cortex, it was not replicable in all animals, and it had a strong correlation with large arteries and veins. After confirming the three major regions of the DMN, we used a neuronal tracing database 3 to investigate their underlying anatomical connections. The three regions had afferent and efferent direct (monosynaptic) anatomical connections with each other (Fig.2), while the mPFC (A32) did not. Based on ultra-high resolution ex-vivo diffusion MRI, we reconstructed two tracts, one was the cingulum that connects the dlPFC with the PCRSC, and the other connects the dlPFC with the PPC and is likely homologous to the superior longitudinal fasciculus of humans (Fig.3). Although the three regions showed strong functional connectivity correlations with direct anatomical connections, they showed different activities to a functional visual task: the posterior regions (PCRSC and PPC) were significantly deactivated by the visual task, but the dlPFC was not (Fig.4), suggesting they may have different functions.DISCUSSION
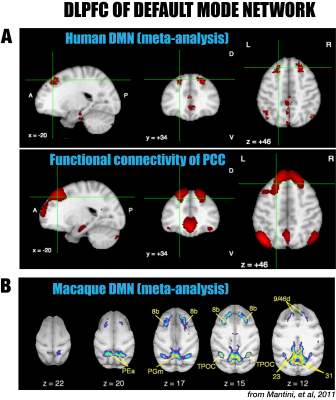
In spite of the strong similarities with the DMN of humans and macaques, the major difference of the marmoset DMN is that the anterior region consists of the dlPFC (8aD and 6DR), but NOT the mPFC. The dlPFC region is more commonly associated with the functions of attention, working memory, and decision making, but is not usually evoked as belonging to the DMN, in spite of its existence in both New World and Old World primates 5-6. However, there is significant evidence in the literature for the inclusion of the dlPFC as a DMN region in both macaques 5 (including areas 8b and 9/46d) as well as in humans 6 (around both middle and superior frontal gyri; Fig. 5). On the contrary, the mPFC, a frontal hub of the DMN of humans, was not a major region in the marmoset DMN. The comparative evidence suggests that the dlPFC, as well as the posterior DMN (PCRSC and PPC), are more conserve evolutionary than the mPFC. Further investigation into the specific role of the dlPFC as a component of the DMN is warranted.Acknowledgements
We thank Xianfeng (Lisa) Zhang, and Dr. Sang-Ho Choi for their help with animal and brain sample preparation; Brandon Chen and Madeline Marcelle for their help with animal training; Dr. Chia-Chun Hung for the visual task fMRI data 1; Dr. Soo Hyun Park for her help with visual task fMRI data and valuable discussions; Dr. Hanbing Lu for providing reverseEPI sequence. Also, many thanks go to the Scientific and Statistical Computing Core of the NIMH Intramural Research Program for their support of AFNI, the High-Performance Computing staff for their support of the NIH-HPC computing clusters, and the valuable databases from the Marmoset Brain Architecture Project and NeuroSynth. This research was supported by the Intramural Research Program of the NIH, NINDS.References
[1] Hung, Chia-Chun, et al. Functional MRI of visual responses in the awake, behaving marmoset. Neuroimage 120 (2015): 1-11.
[2] Cirong Liu, et al, A digital 3D atlas of the marmoset brain based on multi-modal MRI, Under-Review
[3] Majka, Piotr, et al. Towards a comprehensive atlas of cortical connections in a primate brain: Mapping tracer injection studies of the common marmoset into a reference digital template. Journal of Comparative Neurology 524.11 (2016): 2161-2181.
[4] Paxinos, George, et al. The marmoset brain in stereotaxic coordinates. Elsevier Academic Press, 2012.
[5] Mantini, Dante, et al. Default mode of brain function in monkeys. Journal of Neuroscience 31.36 (2011): 12954-12962.
[6] Yarkoni, Tal, et al. "Frontiers: NeuroSynth: a new platform for large-scale automated synthesis of human functional neuroimaging data." Frontiers in Neuroinformatics.
Figures