3261
Bayesian Convolutional Neural Network Based Nonhuman Primate Brain Extraction in Fully Three-dimensional Context1Department of Medical Physics, University of Wisconsin - Madison, Madison, WI, United States, 2Department of Radiology, University of Wisconsin - Madison, Madison, WI, United States, 3Department of Psychiatry, University of Wisconsin - Madison, Madison, WI, United States, 4Department of Biomedical Engineering, University of Wisconsin - Madison, Madison, WI, United States
Synopsis
Brain extraction of MR images is an essential step in neuroimaging, but current brain extraction methods are often far from satisfactory on nonhuman primates. To overcome this challenge, we propose a fully-automated brain extraction framework combining deep Bayesian convolutional neural network and fully connected three-dimensional conditional random field. It is not only able to perform accurate brain extraction in a fully three-dimensional context, but also capable of generating uncertainty on each prediction. The proposed method outperforms six popular methods on a 100-subject dataset, and a better performance was verified by different metrics and statistical tests (Bonferroni corrected p-values<10-4).
Introduction
Brain extraction or skull stripping of magnetic resonance images (MRI) is an essential step in neuroimaging studies, whose accuracy can severely affect subsequent image processing procedures1. Current automatic brain extraction methods demonstrate good results on human brains, but are often far from satisfactory on nonhuman primates2, which are a necessary part of neuroscience research. To overcome the challenges in nonhuman primate brain extraction, we propose a fully-automated brain extraction framework combining deep Bayesian convolutional neural network (CNN) and fully connected three-dimensional conditional random field (CRF), and demonstrate its accuracy, efficiency and flexibility.Methods
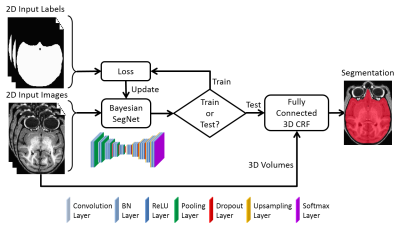
Figure 1 shows the whole framework of the proposed method. The deep Bayesian CNN, Bayesian SegNet3, is used as the core segmentation engine. As a probabilistic network, it is not only able to perform accurate high-resolution pixel-wise brain segmentation, but also capable of measuring the model uncertainty by Monte Carlo dropout testing with dropout sampling at test time4. Then, fully connected three-dimensional CRF is used to refine the probabilistic results from Bayesian SegNet in the fully three-dimensional context of the brain volume5. The proposed method was evaluated in the manner of 2-fold cross validation with a manually brain-extracted dataset comprising 100 rhesus macaque T1w brain volumes, which were collected in a 3T MRI scanner (MR750, GE Healthcare, Waukesha, USA). Our method was also compared with six popular publicly available brain extraction methods on Dice coefficient and average symmetric surface distance2.Results
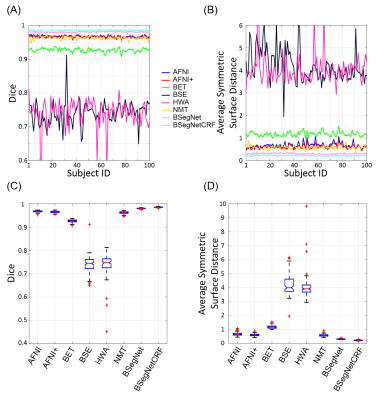
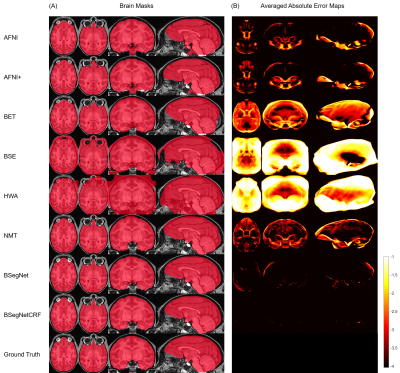
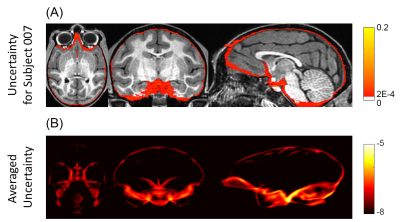
Figure 2(A-B) show that the performance of the proposed method is the best among all the compared methods on each individual’s brain extraction. Figure 2(C-D) show the corresponding boxplots, in which the proposed method outperforms all the compared method with a mean Dice coefficient of 0.985 and a mean average symmetric surface distance of 0.220 mm. The statistical significance of the proposed method was tested against all the other methods with the pairwise Wilcoxon signed rank test (two sided) on both metrics. After Bonferroni correction, all the p-values are still much smaller than 10-4. Figure 3(A) shows the extracted brain masks from all the compared methods for a representative subject, and the proposed method is closest to the ground truth. In Figure 3(B)’s averaged absolute error map in the template space, the proposed method has the best systematic performance with the smallest error distribution and the systematic performance improvement by fully connected three-dimensional CRF can be seen between BSegNet and BSegNetCRF. Figure 4(A) shows the voxel-wise labeling uncertainty of Bayesian SegNet on the representative subject, and Figure 4(B) shows the systematic uncertainty distribution averaged over all the subjects in the template space. Overall, the uncertainty of the brain extraction is very low, and the relative high uncertainty region is at boundary of the brain. Results also show that the uncertainty increases as the training set size decreases or the number of inconsistent labels increases, which matches the expectation well. With an optimized GPU implementation, the prediction time of the whole pipeline is around 2 minutes.Discussion
A new fully-automated brain extraction method is proposed as a combination of deep Bayesian neural network and fully connected three-dimensional CRF for the challenging task of brain extraction in nonhuman primates. Being different from other deep learning based neural networks applied on brain extraction, Bayesian neural network is able to provide the uncertainty of the network on each prediction as well as to predict accurate labels for all the pixels. It is important for a predictive system to generate model uncertainty as a part of the output, since meaningful uncertainty measurement is important for decision-making, especially in medical applications where correct decisions are vital. The combination of fully connected three-dimensional CRF takes the predicted probability maps from Bayesian neural network and moves forward to predictions in a fully three-dimensional context. Because of the limitation of current GPU memory and the huge data size of brain images, it is currently challenging to implement fully three-dimensional brain segmentation solely through deep learning based methods on a single GPU. Thus, with fully connected three-dimensional CRF a complete three-dimensional prediction can be achieved taking into account both the predicted probability maps from deep learning and the image information from the whole original brain volume.Conclusion
In this study, we propose a fully-automated deep learning based brain extraction method on nonhuman primates. The improvement of accuracy by involving fully connected three-dimensional CRF and ability of generating uncertainty for each prediction by utilizing Bayesian neural network are illustrated.Acknowledgements
The authors gratefully acknowledge the technical expertise of Ms. Maria Jesson and Dr. Andrew Fox (UC-Davis), and the assistance of the staffs at the Harlow Center for Biological Psychology, the Lane Neuroimaging Laboratory at the Health Emotions Research Institute, Waisman Laboratory for Brain Imaging and Behavior, and the Wisconsin National Primate Research Center. This work was supported by grants from the National Institutes of Health: P51-OD011106; R01-MH046729; R01-MH081884; P50-MH100031. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
1. Roy, S., Butman, J. A. & Pham, D. L. Robust skull stripping using multiple MR image contrasts insensitive to pathology. NeuroImage 146, 132–147 (2017).
2. Wang, Y. et al. Knowledge-Guided Robust MRI Brain Extraction for Diverse Large-Scale Neuroimaging Studies on Humans and Non-Human Primates. PLoS ONE 9, (2014).
3. Kendall, A., Badrinarayanan, V. & Cipolla, R. Bayesian SegNet: Model Uncertainty in Deep Convolutional Encoder-Decoder Architectures for Scene Understanding. ArXiv151102680 Cs (2015).
4. Gal, Y. & Ghahramani, Z. Bayesian Convolutional Neural Networks with Bernoulli Approximate Variational Inference. ArXiv150602158 Cs Stat (2015).
5. Krähenbühl, P. & Koltun, V. Efficient Inference in Fully Connected CRFs with Gaussian Edge Potentials. ArXiv12105644 Cs (2012).
Figures