3260
A 3D Convolutional Neural Network for Hippocampal Volume Estimation1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Centre Hospitalier Universitaire Vaudois (CHUV), Lausanne, Switzerland, 3Signal Processing Laboratory (LTS 5), École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4CoVii Ltd, Porto, Portugal
Synopsis
Accurate estimation of hippocampal volume is essential for exploiting its sensitivity to pathological changes caused by Alzheimer’s disease (AD) and other forms of dementia. We built and trained a 3D convolutional neural network for fast and accurate segmentation of the hippocampus in T1-weighted structural MR images of the brain. Compared to two software packages (MorphoBox prototype and FreeSurfer), we achieved good disease classification results based on estimated hippocampal volume in a significantly shorter amount of time.
Introduction
Hippocampal volume is a neuroimaging biomarker that was shown to be highly discriminative for the clinical diagnosis of Alzheimer’s disease (AD) and other forms of dementia1,2. Therefore, several automated methods for its volumetric analysis based on structural MRI images have been proposed in the past3. In recent years, convolutional neural networks (CNNs) have shown to be performant in various fields of computer vision, including image segmentation. In this work, we introduce a CNN-based image processing pipeline for segmenting the hippocampus. The method is validated by comparing it to ground truth segmentations and by assessing its ability to provide discriminative information on AD patients as well as patients with Mild Cognitive Impairment (MCI) versus Normal controls.Material and Methods
We designed the CNN as a 3D adaption of the U-net architecture4,5 which consists of a down-sampling path followed by a corresponding up-sampling in order to retrieve segmentation maps with the same dimensions as the input (see Figure 1). Two separate networks were independently trained for left and right hippocampus using Keras6 and TensorFlow7. Training was performed with stochastic gradient descent and Nesterov momentum (learning rate = 1e-3, momentum = 0.99) and a batch size of three using the Harmonized Protocol (HarP8) database consisting of 131 MPRAGE scans registered in MNI space and corresponding manually segmented hippocampal masks. The data was split into test and training sets according to a 4-fold cross-validation. Input to the CNN was a region of interest (ROI, 64x64x64 voxels) selected by registering a randomly chosen template from HarP to the scan. The corresponding manual segmentation masks served as points of reference for the extraction of the ROI. In order to evaluate the network performance against manual gold standard, we computed the following metrics:
- Fuzzy Dice coefficient9
- Relative Volume Difference (RVD): $$$\normalsize\frac{\mid\text{gold standard volume} - \text{estimated volume}\mid}{\text{gold standard volume} + \text{estimated volume}}$$$
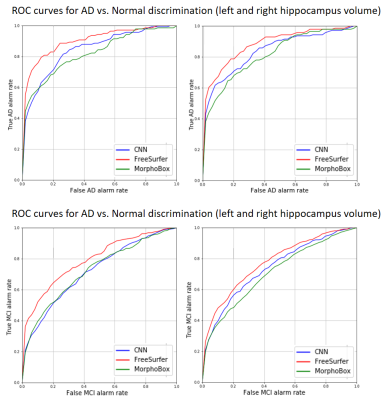
Finally, we compared discriminative power of CNN-estimated volumes to the ones obtained by two volume-based morphometry algorithms: MorphoBox10 prototype and Freesurfer11. This was evaluated on a ADNI standardized12 dataset comprising 673 MPRAGE scans of individuals diagnosed as either normal (N=186), MCI (N=345) or AD (N=142). Several images were removed as they were either already part of the training, corrupted or FreeSurfer failed during processing for an unknown reason. For all algorithms, the resulting hippocampus volumes were normalized by the total intracranial volume obtained by either one of the two morphometry algorithms, and a linear regression against age on the healthy cohort was performed to define a method-specific reference volume range. Discriminative reliability for both AD and MCI cohorts was determined using receiver operating characteristics (ROC) analysis.
Results
Using a GPU (nVidia 1060 GTX mobile), the whole CNN pipeline took approx. 10 seconds to segment both left and right hippocampus while training of the network took ~60 minutes (MorphoBox ~1:30 min, FreeSurfer~10 h). CNN-hippocampus masks were in good agreement with the ground truth according to a mean Fuzzy Dice coefficient of 85.0%$$$\pm$$$1.8% for the left hippocampus and 84.1%$$$\pm$$$2% for the right. As shown in Figure 2, the CNN tended to consistently estimate left and right hippocampus volumes with little spread and a few outliers, as also reflected by mean RVD values of 7.4% $$$\pm$$$ 3.8% and 5.9% $$$\pm$$$ 4.2%, respectively. ROC analysis revealed that CNN discriminated AD/MCI from normal controls with equal sensitivity and specificity of 77.0/65.4 and 73.8/67.7 using left and right hippocampus volume, respectively. As summarized in Table 1, the CNN achieved higher accuracy than MorphoBox whereas Freesurfer outperformed the other two.Discussion and Conclusion
Overall, our CNN-based algorithm accurately estimated hippocampus volumes both quantitatively and qualitatively (see Figure 4) in a timely manner. All methods could detect more than 72% AD patients and close to 63% MCI patients with equal specificity using hippocampus volume estimates. Interestingly, abnormality detection rates obtained using the CNN, MorphoBox and FreeSurfer turned out remarkably consistent on standardized ADNI data despite the different degrees of sophistication and computation time of the respective methods. Possibly, the training set used for the CNN is not representative of the test set. On the other hand, this might be due to a bias caused by an influence on sub-structure volumes due to the global brain atlas registration combined with local intensity distribution modeling that Freesurfer performs. This might be better suited for the hippocampus where gray and white matter are mixed. In conclusion, we have demonstrated the capability of a deep learning architecture to provide fast and accurate segmentations while being trained with relatively little data. Future work should expand the validation on other datasets and further optimize the network.Acknowledgements
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this abstract. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdfReferences
1: Fennema-Notestine C, Hagler DJ, McEvoy LK, et al., Structural MRI Biomarkers for Preclinical and Mild Alzheimer’s Disease. Human brain mapping. 2009;30(10):3238-3253. doi:10.1002/hbm.20744.
2: Niessen, Wiro J., MR brain image analysis in dementia: From quantitative imaging biomarkers to ageing brain models and imaging genetics, Medical Image Analysis, October 2016, Vol.33, pp.107-113
3: Dill, V., Franco, A.R. & Pinho, Automated Methods for Hippocampus Segmentation: the Evolution and a Review of the State of the Art, M.S. Neuroinform (2015) 13: 133
4: Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation, Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, pp. 234–241. Springer (2015)
5: Y. Chen, B. Shi, Z. Wang, et.al., Hippocampus segmentation through multi-view ensemble ConvNets, 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, 2017, pp. 192-196
6: Francois Chollet et.al., Keras, https://github.com/fchollet/keras (2015)
7: Martín Abadi et al., TensorFlow: Large-scale machine learning on heterogeneous systems, https://www.tensorflow.org (2015)
8: Boccardi, Marina et al., Training labels for hippocampal segmentation based on the EADC-ADNI harmonized hippocampal protocol, Alzheimer's & Dementia: The Journal of the Alzheimer's Association , Volume 11 , Issue 2 , 175 - 183
9: Schmitter, Daniel et al., “An Evaluation of Volume-Based Morphometry for Prediction of Mild Cognitive Impairment and Alzheimer’s Disease.” NeuroImage : Clinical 7 (2015): 7–17. PMC. Web. 3 Nov. 2017.
10: Dale, A.M., Fischl, B., Sereno, M.I., 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 179-194
11: Roche A, Ribes D, Bach-Cuadra M, Krger G. On the convergence of EM-like algorithms for image segmentation using Markov random fields. Medical Image Analysis. 2011 Dec;15(6):830–839.
12: B. Wyman, D. Harvey, K. Crawford, et al. and the Alzheimer's Disease Neuroimaging Initiative. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimer's & Dementia, 2012
Figures