3255
Dual Fully Convolutional Networks for Multiscale Context based Robust MRI Skull Stripping1Medical Imaging Technologies, Siemens Healthineers, Princeton, NJ, United States
Synopsis
Brain Segmentation is a standard preprocessing step for neuroimaging applications, but can however be subject to differences in MR acquisition that can lead to added noise, bias field and / or partial volume effect. To address those protocol differences, we therefore present a generic supervised framework, using consecutively two deep learning networks, to produce a fast and accurate brain extraction aimed at being robust across MR protocol variations. While we only trained our network on Human Connectome Project 3T dataset, we can still achieve state-of-the-art results on1.5T cases from LPBA dataset.
Introduction
Brain Segmentation is a standard preprocessing step for neuroimaging applications, often used as a prerequisite for tissue segmentation, and morphometry applications. However, automating this task is challenging, considering the sheer amount of variations of brain shape and size. Protocol differences in MR acquisition can also lead to added noise, bias field and / or partial volume effect. Although brain extraction has traditionally been handled by edge-based, region-based or atlas based methods, each with their advantages and drawbacks, the emergence of machine learning and deep learning in particular, has revived interest in such task, with the main goal of being robust across protocol variation. We therefore present a generic supervised framework, using consecutively two deep learning networks, to produce a fast and accurate brain extraction aimed at being robust across MR protocol variations, while being trained on one dataset.Methods
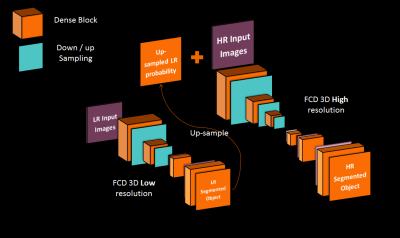
Our supervised framework is in fact, a 3D Dual Network architecture, composed of two Fully Convolutional Dense Networks1 shown in figure 1, each operating at different image resolution. Each network has 12 initial feature maps, a growth rate of 12 and 3 convolutional layers per dense block. The low resolution network (LRN) has 2 levels of pooling and focuses on context (global information about the brain shape and size) while the high resolution network (HRN) has 3 levels of pooling and focuses on recovering details at the brain edge. The LRN network operates at a 2mm isotropic resolution and takes a down-sampled version of the original input to produce low-resolution (LR) segmentation. The probability output of the Softmax activation function is used as context for the next network. It is up-sampled to a high resolution (HR) (1mm isotropic) and concatenated to the HR original input as an additional channel. The brain soft margins of the probability map leave more expression power to the HR network than a binary segmentation with rigid margins would. The final HR segmentation is re-sampled into the original resolution. We trained the networks with a batch-size of 1, using ADAM optimizer and a learning rate of 0.01, driven by a categorical cross-entropy loss. This quite simple process has already been evoked in other ways2-4 but this exact architecture and process is new. Training of the networks occur one after the other. We first pre-train the LR network for getting a good enough context prediction. Then, we use the LR network in a feed-forward fashion to have our HR network contextual input, and train the second network. Prediction at test time is simply a successive feed forward pass in both networks.Results
We used 960 Human Connectome Project (HCP) data5 for our experience, each acquired at 3T, with a 0.7mm isotropic resolution. 800 volumes were used for training and 160 for validation. We trained and tested over resampled data with a 1mm isotropic resolution, but our final test scores are in the original resolution after final resampling. We used the Lanczos algorithm for those resolution changes. Dice coefficients between our prediction and the ground truth were used to evaluate our results. Overall, we obtained an average dice coefficient of 98.6 +/- 0.6, with a specificity of 99.7%+/-0.4 and sensitivity of 98.4%+/-0.1 on our validation sample, as shown in figure 2. In addition, we created a test set with the LPBA dataset, composed by 40 T1w images acquired at 1.5T, with 0.85 x 1.5 x 0.85 resolution. On this test set, Dice coefficient was averaged at 97.5+/-0.4, with sensitivity of 98.2%+/-0.4 and specificity f 99.5% +/-0.2. Figure 3 illustrates our results compared to other Skull Stripping methods tested over LPBA dataset.Discussion
As it has been evoked previously, maximizing the generalization power of a deep learning architecture is one of the main research axes currently. While protocol differences are not specifically addressed here, combining contextual information with a refinement approach via multi-scale Fully Convolutional Dense Net architecture obtains very promising results in early stages of training. Although our model was trained on 3T high resolution data, we can achieve state of the art results compared to other methods that were actually trained on LPBA. Dice, sensitivity and specificity are slightly lower than those of the validation set. Adding more variability to our training set could help improve our predictions.Conclusion
The medical images environment is sparse in terms of image quantity and quality. This architecture allows us to deal with unseen data even in different resolution from the trained data. Then we would be able to use the same model in different institutions.Acknowledgements
No acknowledgement found.References
[1] Jegou S, et al. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. arXiv:1611.09326.
[2] Dong N, et al. Medical image synthesis with context-aware generative adversarial networks. CoRR abs/1612.05362 (2016).
[3] Seyed SMS, et al. Auto-context convolutional neural network for geometry-independent brain extraction in magnetic resonance imaging. CoRR abs/1703.02083 (2017).
[4] Iasonas K. In: Ubernet: Training a ’universal’ convolutional neural network for low-, mid- , and high-level vision using diverse datasets and limited memory. CoRR abs/1609.02132 (2016).
[5] David C. Van Essen, Stephen M. Smith, Deanna M. Barch, Timothy E.J. Behrens, Essa Yacoub, Kamil Ugurbil, for the WU-Minn HCP Consortium. (2013). The WU-Minn Human Connectome Project: An overview. NeuroImage 80(2013):62-79.
Figures