3248
Regularized k-means clustering for segmentation of brain tissues using hemodynamic features in DSC-MRI1Department of Radiation Physics, University of Gothenburg, Gothenburg, Sweden, 2Department of Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden, 3Department of Electical Engineering, Chalmers University of Technology, Gothenburg, Sweden
Synopsis
Inclusion of voxels containing CSF and/or blood vessels can bias ROI statistics used in DSC-MRI analysis. In order to address this problem we propose an automatic method for tissue segmentation based on hemodynamic features obtained from DSC-MRI data. Application of the method in test subjects shows promising results.
Introduction
Dynamic susceptibility contrast (DSC) MRI can be used to characterize the microvasculature of the brain. A commonly used analysis approach is to extract a summary statistic from a region of interest (ROI) in the brain. However, if voxels containing other tissue types such as cerebrospinal fluid (CSF) or blood vessels are included in the ROI, the statistic will be biased. A segmentation that identifies voxels of unwanted tissue types would thus be beneficial. Furthermore, the segmentation should ideally be based on images already included in the examination to reduce examination time.
A segmentation method using the DSC data itself for identification of CSF voxels has been proposed by Kao et al1. In this study, that method is extended to also identify vessels by addition of another feature extracted from the DSC data in combination with regularized clustering.
Methods
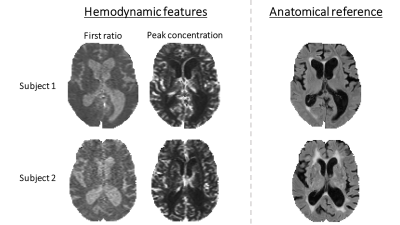
The method for identification of CSF voxels proposed by Kao et al. exploits the long T1 of CSF and the fact that steady state has not been reached during the first dynamics1. The feature, called “first ratio” (FR), is the signal at the first dynamic normalized to some reference. In this study, FR was calculated as
$$FR=\frac{S(1)}{S_0}$$
where S(t) is the signal at the tth dynamic and S0 is the average signal at steady state before contrast agent injection, i.e. during baseline.
Vessels can be assumed to differentiate from other tissue types by having a higher increase in contrast agent concentration after injection. To capture this characteristic, a feature based on the peak concentration (PC) is proposed, calculated as
$$PC=\frac{1}{3} \sum^{T_p+1}_{t=T_p-1}C(t)$$
where C(t) is the contrast agent concentration at the tth dynamic and Tp is the dynamic with maximum C(t), i.e. the peak concentration. Conversion from signal to concentration was performed according to
$$C(t)=-\frac{1}{TE}\ln\frac{S(t)}{S_0}$$
where TE is the echo time.
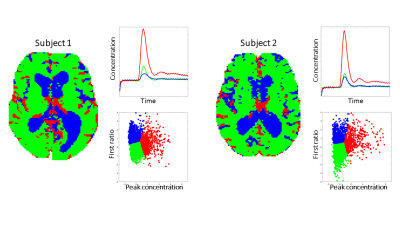
The k-means algorithm2 was used to cluster voxels into three clusters corresponding to CSF, vessels and brain parenchyma. To improve interpretability and robustness of the clustering, a regularization term was added to the k-means objective function
$$J(\mu_k)=\sum_k\left(\sum_{x\in A_k}||x-\mu_k||^2+\lambda_k||\mu_k-\mu_{0,k}||^2\right)$$
where x is the feature vector of a voxel, Ak is the set of voxels in the kth cluster, μk is the center of the kth cluster and μ0,k is the center of the kth cluster proposed prior to the optimization. λk are regularization factors calculated as
$$\lambda_k=\frac{\sum_k\sum_{x\in A_{0,k}}1}{\sum_k\sum_{x\in A_k}1} $$
where A0,k is the set of voxels in the kth cluster proposed prior to the optimization.
A0,k and μ0,k were estimated separately for the CSF and vessel classes. For the CSF prior class, Otsu thresholding3 of the FR feature was used1. Furthermore, connected components (4-connectivity) containing fewer than 10 voxels were excluded from the class. For the vessel prior class the 10 % voxels with highest PC value was chosen. Voxels assigned to both the CSF and the vessel prior classes were excluded. Voxels not included in neither the CSF nor the vessel class were assigned to the brain parenchyma class followed by the same connected component reduction step as for the CSF class. Some voxels were thus not necessarily included in any of the prior sets A0,k. All processing was done using MATLAB.
To study the usefulness of the proposed method, MR images of two idiopathic normal pressure hydrocephalus patients were acquired with a 1.5T Philips Gyroscan Intera. For anatomical reference FLAIR images were acquired (voxel size 1.2×1.2×3mm3). DSC-MR images were obtained every 1000ms with GE-EPI (voxel size 1.8×1.8×5mm3). A bolus (5ml/s) of 0.1mmol/kg body weight Gd-DTPA (Schering, Magnevist 0.5mmol/ml) was initiated at the tenth dynamic into the right antecubital vein. Skullstripping was employed to form an ROI encompassing the brain. For each subject, all voxels within the ROI were extracted and clustered using the described method. Before clustering, features were normalized to have zero mean and unit variance.
Results
The features were highly indicative for the corresponding tissue type (Fig. 1). The resulting clustering showed good performance (see class maps, Fig. 2). The clustering algorithm used information from both features to define clusters in the two-dimensional feature space (see scatter plots, Fig. 2). An appreciable difference between the mean concentration time curves of the different classes was seen, especially when comparing the vessel class with the other classes (see concentration time curves, Fig. 2).Conclusion
This study shows that regularized k-means clustering of brain tissues using two hemodynamic features from DSC-MRI yields promising results with good quality of the resulting segmentation. The proposed method can potentially be used to enhance the quality of ROI based DSC analysis without the need for additional scans.Acknowledgements
The study was supported by grants from the Swedish state under the LUA/ALF agreement.References
- Kao YH, Teng MMH, Zheng WY, Chang FC, Chen YF. Removal of CSF pixels on brain MR perfusion images using first several images and Otsu’s thresholding technique. Magn Reson Med 2010;64:743–748.
- Macqueen J. Some methods for classification and analysis of multivariate observations. Proc Fifth Berkeley Symp Math Stat Probab 1967;1:281–297.
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 1979;9:62–66.
Figures