3247
Multicentric test-retest reproducibility of human hippocampal volumes: FreeSurfer 6.0 longitudinal stream applied to 3D T1, 3D FLAIR and high-resolution 2D T2 structural neuroimaging1Physics and Geology Department, University of Perugia, Perugia, Italy, 2Medical Physics Department, Santa Maria della Misericordia Hospital, Perugia, Italy, 3Division of Neurology V/Neuropathology, Fondazione IRCCS Istituto Neurologico "Carlo Besta", Milan, Italy, 4Laboratory of Alzheimer's Neuroimaging and Epidemiology, IRCCS Fatebenefratelli, Brescia, Italy, 5Memory Clinic and LANVIE-Laboratory of Neuroimaging of Aging, University Hospitals and University of Geneva, Geneva, Switzerland, 6Fondazione IRCCS Istituto Neurologico "Carlo Besta", Milan, Italy, 7Neuroimaging laboratory, IRCCS Santa Lucia Foundation, Rome, Italy, 8Vita-Salute San Raffaele University, Milan, Italy, 9Division of Neuroscience, San Raffaele Scientific Institute, Milan, Italy, 10University of Brescia, Brescia, Italy, 11Department of Neuroscience, Imaging and Clinical Sciences, University “G. d’Annunzio” of Chieti, Chieti, Italy, 12Institute for Advanced Biomedical Technologies (ITAB), University “G. d’Annunzio” of Chieti, Chieti, Italy, 13Centre for Memory Disturbances, Lab of Clinical Neurochemistry, University of Perugia, Perugia, Italy, 14Department of Health Professions - U.O.c Research and Development, Careggi University Hospital, Florence, Italy, 15Department of Neuroradiology, Careggi University Hospital, Florence, Italy, 16Department of Molecular and Translational Medicine, University of Brescia, Brescia, Italy, 17Neuroradiology Unit, University of Brescia, Brescia, Italy, 18Medical Physics Unit, Spedali Civili di Brescia, Brescia, Italy, 19Scientific Direction, Fondazione IRCCS Istituto Neurologico "Carlo Besta", Milan, Italy, 20CIMEC - Center for Mind/Brain Sciences, University of Trento, Trento, Italy
Synopsis
This study evaluates across-session test-retest reproducibility of automatic hippocampus subfields segmentation. A customized acquisition protocol was designed to enhance segmentation reliability in FreeSurfer 6.0 longitudinal analysis stream. Images were processed performing a within-session T1 averaging, using FLAIR images for PIAL surface reconstruction and a high-resolution T2 for the hippocampal subfield segmentation. Results on 12 healthy subjects suggest high reproducibility for different hippocampal subfields and whole hippocampus, generally better than those achievable without T1 averaging and without using FLAIR and high-resolution T2 images.
Introduction
The Italian AD-NET project is a multicentric initiative focused on the development of operational research criteria for early recognition of typical and atypical forms of AD integrating clinical, imaging, and molecular data. As a preliminary part of our Italian AD-NET project, this study reports multicentric test-retest reproducibility of hippocampus and its subfields (Freesurfer 6.0) using the longitudinal processing with FLAIR and high-resolution T2 data for improving pial and subfield segmentation1,2.Methods
Five 3T and one 1.5T clinical MRI sites across Italy currently participate in the project (Table 1). The acquisition protocol (≈40 min in total) included a 3D sagittal T1 (FOV 240x240 mm2, 180 slices, voxel size 1x1x1 mm3, TE 3.9 ms, FA 8°, no fat suppression and no averages) at the beginning and another one at the end of the scanning session, a 3D axial FLAIR (FOV 240x240 mm2, 180 slices, voxel size 1x1x1 mm3, TI 1650 ms, fat suppression and 2 averages) and a coronal T2 (FOV 200x200 mm2, 60 slices, voxel size 0.4x0.4x2 mm3, TE 120 ms, no fat suppression and 2 averages).
Each site recruited 5 healthy volunteers who were scanned in two different sessions a week apart. The preliminary results presented in this study are referred to 12 subjects (9 males and 3 females, average age 53 years, range 35-66 years). A visual quality control of each acquired sequence was performed by an expert clinician before the data analysis.
The structural images were processed according to the longitudinal stream of FreeSurfer 6.0. A within-session T1 averaging was performed to improve the reproducibility of the segmentation of hippocampal subfields3. FLAIR images were used in PIAL surface reconstruction. The standard hippocampal subfield stream4 was then executed on the longitudinal analysis output, using the high-resolution T2 to enhance segmentation reliability. Finally, volumetric information of whole hippocampus (Whole_Hp) and hippocampal subfields, as listed below, were generated: hippocampal tail (Hp_tail), subiculum (Sub), CA1, hippocampal fissure (Hp_fiss), presubiculum (Presub), parasubiculum (Parasub), molecular layer (Hp_ML), GC-ML-DG, CA3, CA4, fimbria, HATA.
Segmentations were visually examined before the statistical analysis to exclude major errors. The test-retest reproducibility error (RE) of the segmentation volumes was calculated as the absolute volume differences across sessions relative to their mean. For each segmented volume, the average RE values across subjects was calculated. Moreover, a comparison between the RE of whole hippocampal volume from both the “aseg.stat” file (Hp_Aseg) and the Whole_Hp was done.
Results
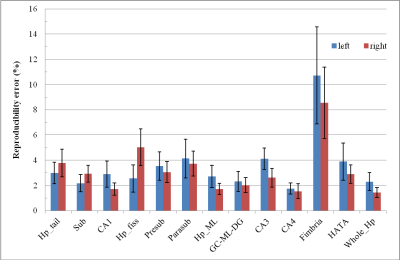
The test-retest average RE for Whole_Hp and hippocampal subfield segmentations are showed in Figure 1, separately for each hemisphere. The reproducibility error is about 2% for right CA1, CA4, right Hp_ML, CG-ML-DG, left Sub and Whole_Hp, whereas it is >5% for Fimbria and right Hp_fiss. These findings are generally better than those of other published studies3 where no FLAIR and high-resolution T2 images were used for the segmentation.
The overall RE (across brain hemispheres average) was 1.1% and 1.9% for Hp_Aseg and Whole_Hp, respectively.
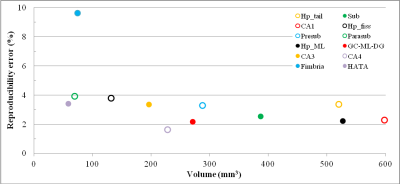
Figure 2 shows hippocampal subfield volumes against the RE. In agreement with previous studies3,5, reproducibility was higher for bigger volumes. However, considering the limited sample size, no correlation between these two variables was calculated. For the same reason, both inter-site and intra-subject right-left variability weren’t estimated.
Conclusions
In this ongoing multi-center study, we evaluated the across-session test-retest reproducibility of hippocampal segmentations on 12 healthy elderly subjects using a customized FreeSurfer stream. Preliminary results suggest high volume reproducibility (RE ≤2%) for the different hippocampal subfields and the whole hippocampus. Importantly, our results show that the use of FLAIR and high-resolution T2 images improves segmentation reproducibility. The next steps include the analysis extension to the whole sample from the consortium for the quantification of within-site and across site reproducibility assessment.Acknowledgements
This study was supported by the Italian Minister of Health, grant number: NET-2011-02346784References
1. http://surfer.nmr.mgh.harvard.edu/fswiki (Accessed November 5, 2017).
2. Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-Subject Template Estimation for Unbiased Longitudinal Image Analysis. Neuroimage, 2012; 61(4):1402–1418.
3. Marizzoni M, Antelmi L, Bosch B et al. Longitudinal reproducibility of automatically segmented hippocampal subfields: a multisite European 3 T study on healthy elderly. Human Brain Mapping, 2015; 36:3516–3527.
4. Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage, 2015; 115:117–137.
5. Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus, 2009; 19:549–557.
Figures