3237
Investigation of Myocardial Fiber Crossings in the Human Heart Using Realistic HARDI Simulation Based on PLI1Key Laboratory of Intelligent Medical Image Analysis and Precise Diagnosis of Guizhou Province, School of Compute Science and Technology, Guizhou University, Guiyang, China, 2School of Computer and Information Technology, Beijing Jiaotong University, Beijing, China, 3Laboratoire TIMC-IMAG, UMR5525 CNRS, Université Grenoble Alpes, Grenoble, France, 4Department of Radiology, Guizhou Provincial People’s Hospital, Guiyang, China, 5Univ.Lyon, INSA-Lyon, Université Claude Bernard Lyon 1, UJM-Saint Etienne, CNRS, Inserm, CREATIS UMR 5220, U1206, F-69621, Lyon, France
Synopsis
We investigate fiber crossings in the myocardium of the human heart using realistic HARDI simulation based on polarized light imaging (PLI). The whole human heart was first imaged using PLI. Based on the fiber orientations measured by PLI, cardiac fiber structures were then modeled, and finally diffusion-weighted images were simulated at different scales using Monte Carlo method and the corresponding ODFs were calculated. The results show that fiber crossings clearly appeared in the myocardium with the increase of scale and that the accuracy of estimating the number of fiber crossings degraded with variable false positive and/or false negative errors.
INTRODUCTION
At the microscopic scale, the myocardium of the human heart is mainly composed of individual myocytes of approximately 25 µm in diameter and 100 µm in length 1. When imaging the whole myocardium of the human heart with current imaging systems that usually have a spatial resolution of millimeter order, we cannot observe the individual myocytes. Instead, we can only observe an aggregate of individual myocytes highly structured and oriented, thus forming the so-called myocardial "fibers" 2. This is the case with popular diffusion tensor imaging (DTI), which can give only a single (averaged) orientation of the oriented structures inside a voxel and for this reason yield fibers that are never crossed. Nevertheless, high angular resolution diffusion imaging (HARDI), largely used for the study of the brain’s microstructures, also appears a potentially useful tool for studying the microstructures of the myocardium. Unfortunately, with the spatial resolution of the millimeter order, current physical HARDI techniques do not allow us to elucidate what the exact fiber complexity of the human myocardium would be. The present study aims to investigate fiber crossings in the human myocardium using realistic HARDI simulation based on polarized light imaging (PLI) that physically measures myocyte orientations with high spatial resolution of the micrometer order.METHODS
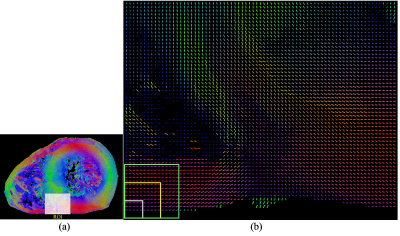
A whole human heart was first imaged by PLI3 with a high spatial resolution of 0.093×0.093×0.5 mm3. Based on the fiber orientations physically measured by PLI (Fig. 1), cardiac fiber structures were then modeled using a cylinder at each voxel of PLI. Finally, diffusion-weighted (DW) images were simulated at different scales using Monte Carlo method and the corresponding orientation distribution functions (ODFs) were calculated. The simulation parameters were: diffusion time=60 ms, pulse duration=2 ms, b-values=5, 700 and 705 s/mm2 for the multi-shell diffusion scheme, number of diffusion sampling directions=192, spatial resolutions=0.465 mm for scale 1, 0.93 mm for scale 2 and 1.395 mm for scale 3, slice thickness=1 mm. The ODFs were reconstructed using the method in the work of Descoteaux4.
Fiber crossings were quantitatively analyzed
at five pixels corresponding to different zones of the myocardium, and this for
each of the three scales. The estimation accuracy is assessed in terms of the
error of estimating the number of fiber crossings. If the estimated number of
crossing fibers is greater than the true one, we call it the false positive
error. Likewise, if the estimated number of fiber crossings is smaller than the
true one, we call it the false negative error.
RESULTS
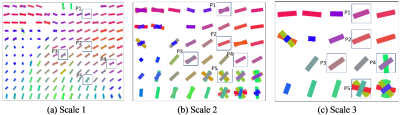
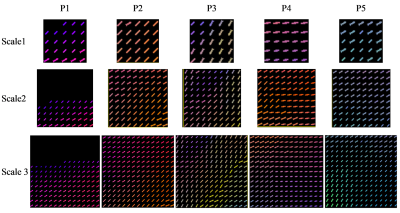
The fiber orientations and crossings derived from ODFs calculated from the simulated DW images at three different scales are shown in Fig. 2. Fiber crossings in the myocardium clearly appeared with the increase of scale. This can readily be seen in Fig. 3. At the smallest scale (scale 1), the orientations of the fibers are rather homogeneous and as a result fiber crossings were not observed at scale 1 (Fig. 2a). On the contrary, at scales 2 and 3, fiber crossings were observed at the pixels in which the fibers have obviously different orientations. This is the case at the pixel P5 for scale 2 and at the pixels P4 and P5 for scale 3.
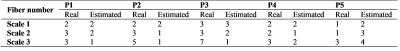
The real and estimated numbers of crossing fibers are given in Table 1. The number of crossing fibers varies from 1 (one fiber, no fiber crossing) to 7 (crossing of 7 fibers, 7-fiber crossings). At scale 1, the number of crossing fibers has been correctly estimated in 80% of cases, and the estimation error is false positive. However, at scales 2 and 3, the number of fiber crossings has been systematically incorrectly estimated; most of the errors are false negative, except at the pixel P5 where the error is always false positive.
DISCUSSION
The revealed myocardial fiber crossings do not imply that individual myocytes are physically crossed. Indeed, the cylinders we put on each voxel in the cardiac structure model were separated and did not touch each other. Therefore, the observed fiber crossings are linked only to imaging scale and reflect the complexity of myocyte orientations in the human myocardium. That is all the true since in the HARDI simulation, we have excluded the influence of noise and fixed other factors such as the acquisition parameters and ODF calculation method.CONCLUSION
Fiber crossings do occur in cardiac HARDI, but depends on the imaging scale or voxel size. Moreover, the number of fibers involved in the crossings is variable, depending on the orientation complexity of fibers inside the voxel.
Acknowledgements
This work was supported by the National Nature Science Foundations of China (Grant No. 61661010, 61671049), the Funds for Talents of Guizhou University (No. 2013(33)), the Nature Science Foundation of Guizhou province (Qiankehe J No.20152044, 20157115, 20161069) and the French ANR (MOSIFAH ANR-13-MONU-0009-01).References
1. Weber KT. Cardiac interstitium in health and disease: The fibrillar collagen network. J Am Coll Cardiol. 1989;13(7):1637-1652.
2. Sun C, Wang L, Yang F, Zhu YBT-I. Investigation of Intravoxel Fiber Configuration Complexity in the Human Heart. In: ISMRM, 2016.
3. Jouk PS, Mourad A, Milisic V, et al. Analysis of the fiber architecture of the heart by quantitative polarized light microscopy. Accuracy, limitations and contribution to the study of the fiber architecture of the ventricles during fetal and neonatal life. Eur J cardio-thoracic Surg. 2007;31(5):915-921.
4. Descoteaux M, Angelino E, Fitzgibbons S, Deriche R. Regularized, fast, and robust analytical Q-ball imaging. Magn Reson Med. 2007;58(3):497-510.
Figures