3224
Test-retest reliability of graph theoretic metrics in adolescent brains1Department of Radiology & Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2Department of Clinical Science Child and Adolescent Psychiatry, Umeå University, Umeå, Sweden, 3Department of Psychology, Stanford University, Stanford, CA, United States, 4Department of Biomedical Sciences, Florida State University, Tallahassee, FL, United States, 5Department of Psychiatry, University of California San Francisco, San Francisco, CA, United States
Synopsis
Graph theory analysis of structural brain networks derived from diffusion tensor imaging (DTI) has been utilized to study neurological and psychiatric disorders but its reliability remains understudied, especially in the still-developing brain. Repeated DTI scans of adolescents were acquired to assess the test-retest reliability of different weighting schemes of brain networks: fractional anisotropy (FA), streamline count (SC), and binary (B). The test-retest scans were performed at two time intervals: 12 weeks apart and within the same scan session, approximately 30 minutes apart. Results suggest that FA-weighting outperforms the other schemes.
Introduction
MRI connectomics treats the brain as a network of interconnected nodes and edges.1 In structural connectivity analysis, nodes represent grey matter ROIs and edges represent white matter tracts typically acquired from diffusion tensor imaging (DTI) and tractography.2 The network is then quantitatively described using graph theory. This method has been used to study many neurological and psychiatric disorders.3-10
However, there are many methodological variations of this complex analysis,11 ultimately hindering its standardization and wider adoption. Test-retest reliability studies have been conducted to resolve this issue12 but few have examined a critical pipeline decision: edge characterization. The first aim of our analysis was to compare the test-retest reliability of graph metrics derived from networks constructed using different weighting schemes: fractional anisotropy (FA)-weighted, streamline count (SC)-weighted, and binary (B).
Additionally, reliability studies typically use adult samples. It is crucial to characterize reliability in a demographic where the brain is still developing. Thus, our second aim was to investigate the potential impact of adolescent brain development on test-retest reliability.
Methods
Two groups of adolescent volunteers received repeated MRI scans in a 3T GE MR750 scanner. The first group (n=21, 14F, mean ± SD = 16.80 ± 1.09yrs) received scans 12 weeks apart, and the second group (n=23, 13F, mean ± SD = 16.64 ± 1.14yrs) received scans within the same session (~30 minutes). The scan included a standard T1-weighted sequence and a DTI sequence (TR = 7.5s, minimum TE, matrix size = 128x128, FOV = 25.6cm, slice thickness = 2mm, 30 directions at b=1000s/mm2, ASSET acceleration factor = 2, acquisition time = 4min).
Diffusion image processing was done using FSL,13 MATLAB, and Diffusion Toolkit,14 and included eddy current correction, DTI reconstruction, and deterministic whole-brain streamline fiber tractography (FACT15). T1-weighted data was registered to the b0-volume of the DTI dataset and to the MNI template using FLIRT,16,17 allowing AAL atlas application in DTI space to produce 90 network nodes. Weighted connectivity matrices were constructed, using three edge definitions: 1) weighted by average fractional anisotropy (FA) within voxels along streamlines connecting two nodes, 2) streamline count (SC), and 3) a binary definition using a density threshold value of 15%.18
Four graph network measures were calculated, two global and two local:
1. Weighted clustering coefficient
2. Weighted characteristic path length
3. Node strength of the R-caudate10
4. Local connection between the R-caudate and the R-middle frontal gyrus (MFG)10
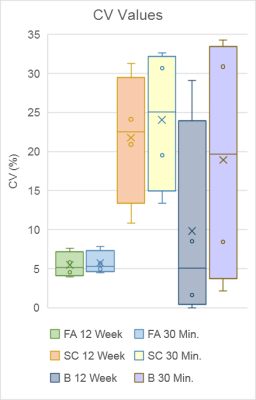
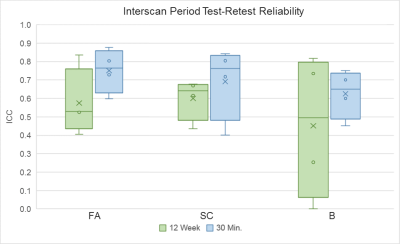
Reliability analysis consisted of: 1) coefficient of variation19,20 (CV) for precision and 2) intraclass correlation coefficient21,22 (ICC) for test-retest reliability. The two combined provide a more nuanced description of reliability. For example, a high ICC and a high CV implies that a measure is sensitive to individual differences and reliable, but is not precise.23,24 See Figure 1 for equations and ICC interpretation.
Results
Figure 2 shows an example of tractograms and brain networks derived from a subject’s within-session DTI scans. Overall, CV values ranged from 0.00 to 34.26 and ICC values ranged from 0.254 to 0.876. FA-weighted ICCs ranged from 0.405 to 0.876. SC-weighted ICCs in total ranged from 0.402 to 0.843. Binary-based ICCs in total ranged from 0.254 to 0.817.Discussion
Regarding our first aim, we found that FA-based graph metrics generally outperformed SC-based ones in the adolescent brain. Both weighted schemes outperformed binary metrics. FA-based measures displayed simultaneous high reliability and precision. SC-based measures showed mixed precision and performed particularly worse than binary measures for global metrics. The superior performance of the FA-weighting may come from its more biological underpinnings, namely its reflection of white matter fiber microstructure.25 Comparatively, SC is a more abstract measure that is swayed by changes in tract length, curvature, and degree of branching.26
For our second aim, we found that weighted schemes produced reliable measurements over 12 weeks despite potential adolescent developmental interference. Again, weighted measures outperformed binary ones, with FA-weighting producing consistent, reliable results in this longitudinal context.
Based on our results, we recommend using weighted over binary edge characterizations, and favoring FA- over SC-based weights. Our findings also indicate that graph network analysis is a feasible method over longer periods of time (i.e. three months), and in situations potentially impacted by neurodevelopmental factors. We also recommend using FA-weighted definitions for this longitudinal context.
Acknowledgements
NCCIH R21AT009173, NICHD R01HD072074, UCSF Research Evaluation and Allocation Committee (REAC) and J. Jacobson Fund, and UCSF Radiology Seed Grant #14-31.References
1. Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10, 186–198 (2009).
2. Rubinov, M. & Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 52, 1059–1069 (2010).
3. He, Y., Chen, Z. & Evans, A. Structural Insights into Aberrant Topological Patterns of Large-Scale Cortical Networks in Alzheimer’s Disease. J. Neurosci. 28, 4756–4766 (2008).
4. Verstraete, E., Veldink, J. H., Mandl, R. C. W., Berg, L. H. van den & Heuvel, M. P. van den. Impaired Structural Motor Connectome in Amyotrophic Lateral Sclerosis. PLOS ONE 6, e24239 (2011).
5. Bernhardt, B. C., Chen, Z., He, Y., Evans, A. C. & Bernasconi, N. Graph-Theoretical Analysis Reveals Disrupted Small-World Organization of Cortical Thickness Correlation Networks in Temporal Lobe Epilepsy. Cereb Cortex 21, 2147–2157 (2011).
6. Caeyenberghs, K. et al. Brain connectivity and postural control in young traumatic brain injury patients: A diffusion MRI based network analysis. NeuroImage: Clinical 1, 106–115 (2012).
7. Bos, D. J. et al. Structural and functional connectivity in children and adolescents with and without attention deficit/hyperactivity disorder. J Child Psychol Psychiatr 58, 810–818 (2017).
8. Leow, A. et al. Impaired Inter-Hemispheric Integration in Bipolar Disorder Revealed with Brain Network Analyses. Biological Psychiatry 73, 183–193 (2013).
9. Korgaonkar, M. S., Fornito, A., Williams, L. M. & Grieve, S. M. Abnormal Structural Networks Characterize Major Depressive Disorder: A Connectome Analysis. Biological Psychiatry 76, 567–574 (2014).
10. Tymofiyeva, O. et al. DTI-based connectome analysis of adolescents with major depressive disorder reveals hypoconnectivity of the right caudate. J Affect Disord 207, 18–25 (2017). Fornito, A., Zalesky, A., Pantelis, C. & Bullmore, E. T. Schizophrenia, neuroimaging and connectomics. NeuroImage 62, 2296–2314 (2012).
11. Meskaldji, D. E. et al. Comparing connectomes across subjects and populations at different scales. Neuroimage 80, 416–425 (2013).
12. Welton, T., Kent, D. A., Auer, D. P. & Dineen, R. A. Reproducibility of Graph-Theoretic Brain Network Metrics: A Systematic Review. Brain Connect 5, 193–202 (2015).
13. Smith, S. M. et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23, S208–S219 (2004).
14. Wang. Diffusion toolkit: a software package for diffusion imaging data processing and tractography.
15. Mori, S., Crain, B. J., Chacko, V. P. & Van Zijl, P. C. M. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 45, 265–269 (1999).
16. Jenkinson, M. & Smith, S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis 5, 143–156 (2001).
17. Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 17, 825–841 (2002).
18. Duda, J. T., Cook, P. A. & Gee, J. C. Reproducibility of graph metrics of human brain structural networks. Front Neuroinform 8, (2014).
19. Lachin, J. M. The role of measurement reliability in clinical trials. Clin Trials 1, 553–566 (2004).
20. Vaessen, M. J. et al. The effect and reproducibility of different clinical DTI gradient sets on small world brain connectivity measures. NeuroImage 51, 1106–1116 (2010).
21. McGraw, K. O., & Wong, S. P. Forming inferences about some intraclass correlation coefficients. Psychological Methods, 1, 30–46 (1996).
22. Shrout, P. E., & Fleiss, J. L. Intraclass correlations: Uses in assessing rater reliability. Psychological Bulletin, 86, 420–428 (1979).
23. Andreotti, J. et al. Repeatability Analysis of Global and Local Metrics of Brain Structural Networks. Brain Connectivity 4, 203–220 (2014).
24. Owen, J. P. et al. Test-retest reliability of computational network measurements derived from the structural connectome of the human brain. Brain Connect 3, 160–176 (2013).
25. Pierpaoli, C. & Basser, P. J. Toward a quantitative assessment of diffusion anisotropy. Magn. Reson. Med. 36, 893–906 (1996).
26. Jones, D. K., Knösche, T. R. & Turner, R. White matter integrity, fiber count, and other fallacies: The do’s and don’ts of diffusion MRI. NeuroImage 73, 239–254 (2013).
27. Cicchetti, D. V. Multiple comparison methods: Establishing guidelines for their valid application in neuropsychological research. Journal of Clinical and Experimental Neuropsychology, 16, 155–161 (1994).
28. Bastian, M., Heymann, S., & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media, 8, 361-362 (2009).
Figures
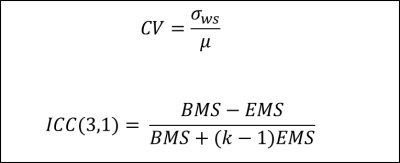
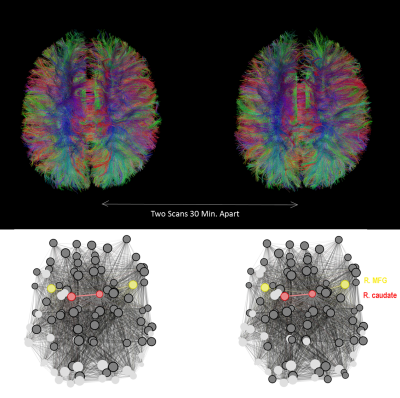
Figure 2. Top) Tractograms derived from a subject’s two DTI scans taken 30 minutes apart (within-session). Bottom) A brain network map of the same subject using 90 nodes derived from the AAL atlas. The nodes representing the R-middle frontal gyrus (R. MFG) are depicted in yellow; the caudate is depicted in red. Nodes connected to the right caudate are shown in dark grey; those that are not connected are shown in light grey. Network visualization was performed using Gephi.28