3189
Sampling order optimization preserves contrast and improves clinical diagnostic utility of accelerated prospective 3D brain MRI: a radiological assessment study on healthy volunteers1School of Engineering, Institute for Digital Communications, University of Edinburgh, Edinburgh, United Kingdom, 2Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
Synopsis
This study shows the importance of sampling order optimization for the contrast preservation of accelerated prospective 3D MRI leading to the improvement in clinical diagnostic utility of accelerated scans using compressed sensing and parallel imaging reconstructions.
Purpose
The main purpose of this study is to validate
the clinical diagnostic utility of accelerated sampling order optimized
prospective MRI in hospitals by radiological assessment of image quality and
artefacts for reducing the overall scan duration. Sampling order optimization
is important because, in certain clinical sequences like Inversion Recovery
(IR) prepared 3D Gradient Echo (GRE), each readout starts at the effective TE
that has optimum contrast. For each line of ky space, a train of 80 kz samples
(with TR=9.6 ms) is acquired after each inversion pulse (with 500 ms delay). Therefore,
the samples collected at the beginning of the readout have better contrast than
those towards the end. Hence, it is important to ensure that the central
kz-space data which contains most of the contrast information in the image is
acquired towards the beginning of each readout for contrast preservation1. In this study, we compare the
radiological scores of sampling order optimized accelerated prospective 3D
scans with the fully sampled scans to determine whether accelerated scans can
be used for clinical diagnosis.Methods
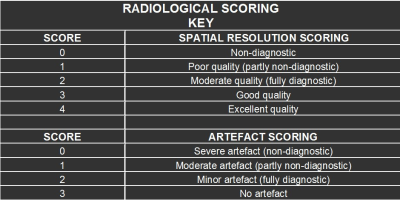
The scanning was performed on a 1.5T GE Signa Horizon HDX scanner using the manufacturer’s 3D IR-prepared GRE sequence on 8 subjects who were recruited under the healthy volunteer ethics protocol. Informed consent was received from the subjects prior to the scanning. Sequence parameters were TR/TE/TI=10/4/500 ms; flip angle = 8°; matrix 192×192×160 slices; isotropic 1.3 mm voxels. Fully sampled and accelerated prospective scans (by using patterns without sampling order optimization – Fig. 1 and with sampling order optimization – Fig. 2) were performed on each volunteer and the images were reconstructed using the compressed sensing-parallel imaging (CS-PI) based NESTA algorithm2. The acquired fully sampled and subsampled brain datasets were then randomised and given to a neuroradiologist for assessment of image quality and artefacts. The radiological scoring key is shown in table 1 in which the brain was divided into 4 separate regions that were scored (i.e. basal nuclei, brainstem, temporal gyri and precentral gyri) with 0 being the lowest score (i.e. non-diagnostic) and 4 being the highest (i.e. excellent quality). Artefact scoring of the image datasets was also done and it ranged from 0 (i.e. severe artefact) to 3 (i.e. no artefact)3.Results
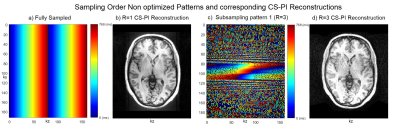
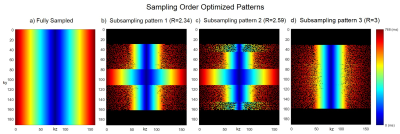
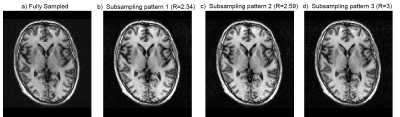
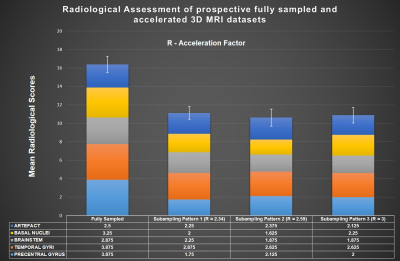
Fig. 1 shows the fully sampled (1a) and three times accelerated subsampling pattern (1c) without sampling order optimization along with its corresponding CS-PI reconstructions (1b and 1d). Fig. 2 shows the sampling order optimized fully sampled (2a) and subsampled patterns (2b, 2c and 2d) at various acceleration factors (R). The colourbars in Fig. 1 and Fig. 2 illustrate the time instant at which the samples are collected during each readout train which starts after an initial inversion pulse of 500 ms duration. Fig. 3 shows the corresponding CS-PI reconstructions of the sampling order optimized fully sampled (3a) and subsampled patterns (3b, 3c and 3d) for a single slice of the 3D brain scan. Fig. 4 shows the mean radiological scores (S) along with standard error (SE) of the fully sampled and subsampled datasets in which each column was further subdivided into the mean radiological scores of the four different brain regions and the mean artefact scores. The CS-PI reconstructions of accelerated scans (up to R= 3) produced images that were fully diagnostic. The scan time was reduced from 8:08 minutes to 2:42 minutes for R= 3.Discussion and Conclusion
Although the fully sampled images routinely had higher radiological scores compared to the accelerated images, it can be seen from Fig. 4 that the accelerated scans were still fully diagnostic (S > 2) except for one or two cases in different brain regions that were partly non-diagnostic (S < 2). This variability may be attributed to motion during the scanning and is more likely to improve with more volunteer scans because each S was closer to being fully diagnostic (i.e. S = 2) than partly non-diagnostic (i.e. S = 1). These results show that sampling order optimization of subsampling patterns improved the clinical utility of accelerated scans by producing clinically usable images up to R = 3. The deep brain grey matter could also be easily identified in sampling order optimized accelerated scans shown in Fig. 3 due to contrast preservation while it was not the case when sampling order optimization was not performed as shown in Fig. 14. Further acceleration (R > 3) produced reconstruction artefacts that made the images clinically unusable. However, these artefacts could be potentially supressed by the use of more sophisticated compressed sensing algorithms based on non-local means5 and a current study is ongoing to validate it.Acknowledgements
The research leading to these results has received funding from the European Union's H2020 Framework Programme (H2020-MSCA-ITN-2014) under grant agreement no 642685 MacSeNetReferences
[1] A. J. V. Benjamin, W. Bano, M. Davies, and I. Marshall, "Sampling Order Optimization for contrast preservation in accelerated prospective 3D MRI," in Proceedings of ESMRMB, 2017, p. #256.
[2] S. Becker, #233, #244, m. Bobin, E. J. Cand, and #232, "NESTA: A Fast and Accurate First-Order Method for Sparse Recovery," SIAM J. Img. Sci., vol. 4, pp. 1-39, 2011.
[3] S. Sirin, S. L. Goericke, B. M. Huening, A. Stein, S. Kinner, U. Felderhoff-Mueser, et al., "Evaluation of 100 brain examinations using a 3 Tesla MR-compatible incubator—safety, handling, and image quality," Neuroradiology, vol. 55, pp. 1241-1249, 2013.
[4] I. Marshall, G. Rilling, Y. H. Tao, C. Du, S. Varma, D. Job, et al., "Radiological and quantitative assessment of Compressed Sensing reconstruction of undersampled 3D brain images," in Proceedings of ISMRM, 2015, p. #2497.
[5] C. Ulas, P. A. Gomez, J. I. Sperl, C. Preibisch, M. I. Menzel, A. Haase, et al., "A Robust Reconstruction Method for Quantitative Perfusion MRI: Application to Brain Dynamic Susceptibility Contrast (DSC) Imaging," in Proceedings of ISMRM, 2017, p. #209.
Figures