3186
Nonlocal multispectral image filtering to improve determination of cerebral blood flow from pseudo-continuous arterial spin labeling imaging1National Institute on Aging, National Institutes of Health, Baltimore, MD, United States
Synopsis
Changes in the cerebral blood flow (CBF), measured using arterial spin labeling (ASL), are an emerging biomarker for normal aging, Alzheimer's disease, and other neurodegenerative conditions. However, ASL signal-to-noise ratio (SNR) is inherently low, diminishing the quality of CBF determination. While attempts have been made to improve SNR in ASL images using post-processing filters, performance is limited and several user-defined parameters are required adding further complexity in implementation. Here, we introduce a simple, novel filtering algorithm and demonstrate its potential to enhance the quality of CBF mapping.
PURPOSE
Arterial spin labeling (ASL) MRI uses arterial blood water as an endogenous tracer to measure cerebral blood flow (CBF). However, the signal-to-noise ratio (SNR) in ASL experiments is inherently low. We present a simple, novel denoising approach to enhance SNR in ASL images and to improve CBF determination.MATERIALS & METHODS
Image filtering
Nonlocal noise-reducation filters replace the signal intensity within a voxel i with an improved estimate of its underlying amplitude, without decreasing image resolution. This requires selection of voxels that represent similar tissue. The recently-introduced NESMA filter (1-3) restores the amplitude of an index voxel based on M pre-selected voxels with similar multispectral signal patterns. The control image, SC, labeled image, SL, and proton density (PD) image, SPD, from which a CBF map can be derived (4), may be regarded as multispectral representations. Then the respective estimates, $$$\widehat{A_{C}}(i)$$$, $$$\widehat{A_{L}}(i)$$$, and $$$\widehat{A_{PD}}(i)$$$ of the true amplitudes in voxel i of these three images are given by:
$$\widehat{A}_{C,L,PD}(i)=M^{-1}\sum_j^M S_{C,L,PD}(j).\ \ \ \ \ \ \ \ \ [1]$$
The goal of the filtering algorithm is to appropriately select the M voxels used for averaging; in our implementation, the similarity between two voxels i and j is calculated using the relative Euclidean distance (RED):
$$RED(i)=100\times\sqrt{\frac{(S_C (i)-S_C (j))^2+(S_L (i)-S_L (j))^2+(S_{PD} (i)-S_{PD} (j))^2}{S_C^2 (i)+S_L^2 (i)+S_{PD}^2 (i)}}.\ \ \ \ \ \ \ \ \ [2] $$
RED is calculated between the index voxel, i, and every other voxel, j, within a large search window, R, centered around i. R is selected to be large enough to permit inclusion of multiple voxels, but small enough so that B1 and noise are approximately constant. The M voxels with the smallest values of RED are then selected. This selection results in a subset of M voxels designated as similar to i from which $$$\widehat{A_{C}}(i)$$$, $$$\widehat{A_{L}}(i)$$$, and $$$\widehat{A_{PD}}(i)$$$ are computed using Eq.1. We denote this filter as NESMA-ASL.
CBF mapping
In each voxel, i, the value of CBF was calculated using (4):
$$CBF(i)=6000\times\frac{λ\ (I_C (i)-I_L (i))\ exp(PLD⁄T_{1,Blood})}{2\ α \ T_{1,Blood}\ I_{PD}(i)\ (1-exp(-τ⁄T_{1,Blood}))}\ \ [mL/100 g/min],\ \ \ \ \ \ \ \ \ [3]$$
where λ is the brain-blood partition coefficient, α is the labeling efficiency, T1,Blood=1.65s is the longitudinal relaxation time of blood, and τ and PLD are, respectively, the labeling and post labeling durations. IC, IL, and IPD are the control, labeled, and PD images, respectively. Other signal models are available; our analysis applies to these as well, and our use of Eq.3 may be regarded as illustrative.
In-vivo analysis
After informed consent, healthy volunteers (male, 24 and 50 years old; female, 57 and 61 years old) were scanned with a 2D pCASL sequence combined with single-shot EPI acquisition on a 3T Philips system, using an 8-channel head coil. For each participant, SC, SL, and SPD images were independently acquired at low resolution (LR): 2.8mm x 2.8mm x 5mm, reconstructed to 1.5mm x 1.5mm x 5mm, and high resolution (HR): 1.9mm x 1.9mm x 5mm, reconstructed to 1mm x 1mm x 5mm. Other experimental parameters were: TR=7.5s, τ=1.8s, PLD=2s, and signal averages=30. CBF maps were derived from unfiltered or filtered ASL images using NESMA-ASL (with RED < 5% and R=11voxels x 11voxels x 1voxel). For comparison, we also applied Gaussian averaging (GA) and boxcar averaging (BA) filters (kernel size=5 voxelsx 5voxels x 1voxel).
RESULTS & DISCUSSION
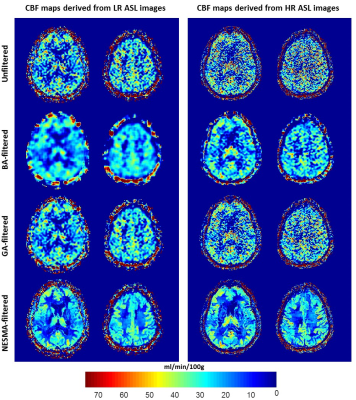
Fig.1 shows substantial random variation in CBF maps derived from unfiltered images. Random variation was reduced using GA or BA, but at the expense of loss of image detail. In contrast, CBF maps derived from images filtered with NESMA-ASL exhibited preservation of detail as well as greatly reduced random variation. Furthermore, while LR and HR CBF maps were, overall, similar in most brain regions, HR mapping allows greater depiction of details of the CBF distribution.
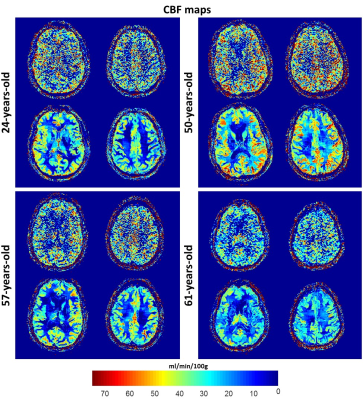
Fig.2 shows a comparison of CBF maps from the brains of the four subjects. CBF maps calculated from HR unfiltered or filtered images using NESMA-ASL are displayed for two different slices. The results indicate higher CBF values in the middle-aged subject in several image regions as compared to the younger or older subjects. These results suggest that an inverted U shape may best describe the relationship between age and CBF, as discussed in (5), and demonstrate the sensitivity of the method to CBF changes with age (6-7).
CONCLUSIONS
The NESMA-ASL filter permitted construction of high-resolution, high-precision CBF maps in the human brain from pCASL imaging data. The use of NESMA-ASL for pCASL as well as other ASL modalities may contribute significantly to the goal of high quality, high resolution CBF mapping.Acknowledgements
This work was supported by the Intramural Research Program of the National Institute on Aging of the National Institutes of Health.References
1. Bouhrara M, Bonny JM, Ashinsky BG, Maring MC, Spencer RG. Noise Estimation and Reduction in Magnetic Resonance Imaging Using a New Multispectral Nonlocal Maximum-likelihood Filter. IEEE transactions on medical imaging. 2017;36(1):181-93.
2. Maring. MC, Bouhrara. M, Spencer RG. A Fast Adaptive Multispectral Nonlocal Denoising Filter. ISMRM; Honolulu, HI, USA 2017.
3. Bouhrara M, Maring MC, Reiter DA, Bonny JM, Spencer RG. Enhanced Quality of Myelin Water Fraction Mapping from GRASE Imaging Data of Human Brain using a New Nonlocal Estimation of multi-Spectral Magnitudes (NESMA) Filter. ISMRM; Honolulu, HI, USA 2017.
4. Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magnetic Resonance in Medicine 2015 ;73 :102-116.
5. Heo S, Prakash RS, Voss MW, Erickson KI, Ouyang C, et al. Resting Hippocampal Blood Flow, Spatial Memory and Aging. Brain Research 2010;1315:119-27.
6. Liu Y, Zhu X, Feinberg D, Guenther M, Gregori J, et al. Arterial spin labeling MRI study of age and gender effects on brain perfusion hemodynamics. Magnetic Resonance in Medicine 2012;68(3):912-22.
7. Rusinek H, Brys M, Glodzik L, Switalski R, Tsui WH, et al. Hippocampal blood flow in normal aging measured with arterial spin labeling at 3T. Magnetic Resonance in Medicine 2011;65(1):128-37.
Figures