3171
Brain Vessel Extraction without MRA / V using Deep Convolutional Neural Network1Electrical and Electronic Engineering, Yonsei University, Seoul, Republic of Korea, 2Philips Korea, Seoul, Republic of Korea, 3Radiology and Research Institute of Radiological Science, Yonsei University College of Medicine, Seoul, Republic of Korea
Synopsis
In this paper, we introduce a deep residual learning approach to extract brain vessels from contrast-enhanced(CE) magnetic resonance images. Our experiment results show that we can successfully achieve and visualize brain vessel information from CE MRI without magnetic resonance angiography and magnetic resonance venography(MRA/V) that are currently used for brain vessel extraction.
Introduction
MRA/V differs from MRI in that it specifically looks at arteries in human body while MRI is used for tissue evaluation such as metastatic lesion detection. Therefore, MRA/V have been required to achieve vessel information for complete diagnosis and this cost additional money and time. Our research started from the idea that vessel extraction from MRI will lead to much efficient diagnosis process, decreasing the scanning time and necessity of additional MRA/V scanning. To the best of our knowledge, no research has focused on extracting brain vessels directly from MRI data without the help of MRA/V data. Most of the conventional vessel extraction algorithms utilize the high intensity signal of MRA/V which makes them inappropriate to apply on MRI1-3. Otherwise, they use MRI only as a prior information to facilitate the vessel segmentation within MRA4. In this study, we implemented a deep convolutional neural network(CNN) with skip connection to extract brain vessels from contrast-enhanced gradient-echo(CE-GRE) sequence data. This approach can furthur be applied to suppressing vessles in CE-GRE data to highlight metastatic lesions.Methods
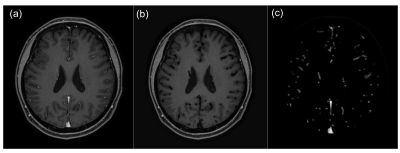
In CE T1-weighted scans, a contrast agent(usually gadolinium) is administered to increase the signal intensity of vessels and surrounding tissues, and thereby discover abnormalities in tissue. We utilized the relatively high intensity of vessels in CE T1-weighted images to extract them. CE T1-weighted images were scanned from a 3.0-T MRI Scanner(Ingenia CX, Philips Medical Systems, Best, The Netherlands) with an eight-channel sensitivity-encoding head coil using 3D spoiled gradient-echo sequence. The image acquisition parameters for were as follows : FOV = 20-24cm; matrix = 240x240; section thickness = 1mm. A total of 5 subject data were available and we selectively used 90 consecutive slices in axial direction from each subject as training data.
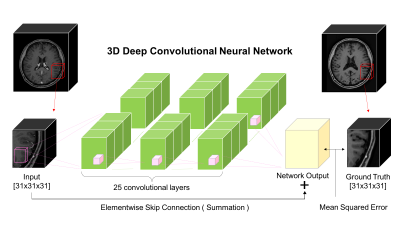
First, brain vessel masks were created using MIPAV software5. We manually segmented vessels on every 2D axial slice and concatenated these to make a 3D vessel mask. Using this mask, brain vessels in the original data were suppressed to zero intensity to obtain images that differ from original images only in vessel region. Next, the original dataset and vessel-suppressed dataset were fed as input and output of our deep CNN so that the network could suppress brain vessels by learning their non-linear features. Our deep CNN is depicted in Figure 1. It capitalizes on its deeply stacked 25 convolutional layers so that the cascaded filters can efficiently exploit contextual information from input data. In addition, the elementwise skip connection between input and output of the last convolutional layer helps the network to converge faster since it helps the network to learn only the residuals, which are brain vessels in our case. This type of architecture can be effectively used when input and output are highly correlated6, which makes it suitable for our vessel extracting problem. Mean squared error between the ground truth and network output was used as a loss function to train the network.
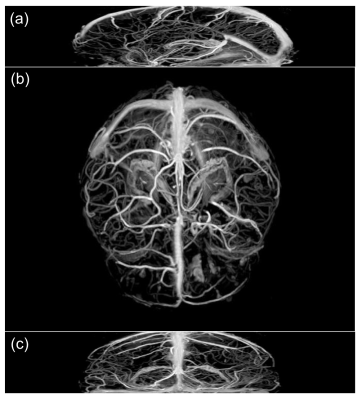
Our network was trained on patches of size 31x31x31. Each convolutional layer consists of kernels of size 3x3x3 with 64 feature maps except the last layer which has only 1 feature map to match the output size with the ground truth. Batch normalization and rectified linear unit(ReLU) activation were performed after each convolutional layer except the last one. Kernel weights were updated by backpropagation using Adam optimizer. Once training is done, our proposed network is capable of suppressing the brain vessels of unseen CE-GRE data to zero intensity. The network output of this test data was then subtracted from the original test data to extract brain vessels(Figure 2). Visualization of the extracted vessels was acquired using maximum intensity projection(MIP) on axial, sagittal, and coronal planes.
Results
Figure 3 shows the performance of proposed method by displaying MIP images of extracted vessels in axial, sagittal, and coronal planes. It shows that proposed method can successfully extract brain vessel information from CE-GRE data. The coverage of the extracted vessels can change according to the range of the slices we use for training.Conclusion
In this paper, we propose a deep residual learning approach to extract brain vessels without MRA/V. Our trained model shows that a deep-learned network can extract detailed structures of brain vessels in CE-GRE MRI data. Since creating training data by manual segmentation takes much time, an automatic algorithm to complement this pre-processing step should be designed to increase the number of training data and improve performance of the proposed method. Future works will focus on applying this method to effectively eliminating brain vessels in CE-GRE images to highlight metastatic lesions.Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government(MSIP) (2016R1A2R4015016).References
1. Vasilevskiy, Alexander, and Kaleem Siddiqi. "Flux maximizing geometric flows." IEEE transactions on pattern analysis and machine intelligence 24.12 (2002): 1565-1578.
2. Eiho, S., et al. "Branch-based region growing method for blood vessel segmentation." Proceedings of International Society for Photogrammetry and Remote Sensing Congress. 2004.
3. Descoteaux, Maxime, D. Louis Collins, and Kaleem Siddiqi. "A geometric flow for segmenting vasculature in proton-density weighted MRI." Medical image analysis 12.4 (2008): 497-513.
4. SARAN, AYŞE NURDAN, Fatih Nar, and Murat Saran. "Vessel segmentation in MRI using a variational image subtraction approach." Turkish Journal of Electrical Engineering & Computer Sciences 22.2 (2014): 499-516.
5. McAuliffe, Matthew J., et al. "Medical image processing, analysis and visualization in clinical research." Computer-Based Medical Systems, 2001. CBMS 2001. Proceedings. 14th IEEE Symposium on. IEEE, 2001.
6. Kim, Jiwon, Jung Kwon Lee, and Kyoung Mu Lee. "Accurate image super-resolution using very deep convolutional networks." Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016.
Figures