3170
Novel Imaging Phantom for Accurate and Robust Measurement of Brain Atrophy Rates Using Clinical MRIHoushang Amiri1, Iman Brouwer1, Joost P.A. Kuijer1, Jan C. de Munck1, Frederik Barkhof1,2, and Hugo Vrenken1
1Radiology & Nuclear Medicine, VU University Medical Center, Amsterdam, Netherlands, 2Institutes of Neurology and Healthcare Engineering, UCL, London, United Kingdom
Synopsis
MR images are widely used to measure brain atrophy in neurodegenerative diseases. However, reliable evaluation of atrophy is hampered by scanner-induced systematic variability. Here, we developed an MR-compatible phantom and analysis software for robust and reliable evaluation of the brain volume loss. The phantom was made using 3D-printing and contains three inflatable brain structures equipped with a precise volume change system. The phantom was imaged at three different clinical 3T MR scanners and images were analyzed by our developed software. This phantom can accurately and robustly provide a selected volume change to mimic a certain disease.
INTRODUCTION
Brain volume loss (atrophy) has been proven to be an important characteristic of neurological diseases such as Alzheimer’s disease (AD). However, scanner-induced systematic differences in atrophy measures have been reported using different methods 1-4. Therefore, to increase statistical power in clinical trials and to establish atrophy measures as reliable clinical biomarkers, such scanner-induced effects should be minimized. In this study we developed and performed initial validation tests of an MR-compatible phantom and analysis software for robust and reliable evaluation of the brain volume loss.METHODS
The MRI phantom contains inflatable structures equipped with a precise volume change system (FIG. 1). The phantom contains three brain structures: a cerebral hemisphere, putamen, and caudate nucleus. The hemisphere model was created by scanning a post-mortem healthy brain using a surface 3D scanner (Artec Spider, Arc 3D). The models of putamen and caudate nucleus were generated from MR images of healthy control subjects using FSL-FIRST. Using these models, molds of all three structures were 3D-printed using ZP151 powder (3D systems). Then, the molds were repeatedly dipped into pre-vulcanized latex rubber (pvs-elastica, FromX), to reach a homogenize thickness. Finally, the molds were dissolved in water and phantoms remained. Syringes (Hamilton®, Switzerland) of 100mL and 100µL were incorporated with a custom-made stainless steel plunger with a finely bored syringe barrel to change the volume in the hemisphere and the small structures, respectively. To validate the method, the phantom was imaged at three different clinical 3T MR scanners (GE, Toshiba and Philips) using a standard clinical 3D gradient echo sequence available at each scanner with voxel volumes 1-1.2 µL (FIG. 2). In each scanning session, 5 volume changes were applied as indicated in FIG. 3a, ranging between about 1-5% of the baseline volume. Next, we developed software to measure volume change from MR images, using existing methods and pipelines (FIG. 4). Then, the measured volume change was compared with the known (applied) volume change using intra-class correlation coefficient (ICC, absolute agreement) and mean absolute difference (MAD). SPSS version 22 (IBM) was used.RESULTS
FIG. 2 shows example phantom MR images. The volume change calculated by our software versus the applied volume change is presented in FIG. 5. ICC ranged from 0.999-1 for hemisphere, 0.976-0.998 for putamen, and 0.985-0.999 for caudate nucleus (FIG. 3b). The MAD were 109-332µL for hemisphere, 2.9-11.9µL for putamen, and 2.2-10.1µL for caudate nucleus (FIG. 3b).DISCUSSION and CONCLUSIONS
Although other anthropomorphic brain phantoms have been described 5-7, to our knowledge this is the first structural MR-compatible phantom aiming to standardize volume change measurements within and between scanners. Our device can accurately and robustly provide a selected volume change to mimic e.g. the change typically observed in a certain disease. The developed software can measure the volume changes from the images with high precision. Next step is to validate this in patient studies, targeting a method to standardize volume change measurements obtained on different scanners.Acknowledgements
We would like to thank kind technical supports by the 3D Innovation Lab at the VU University Medical Center, specially D. Koops, F. Verver and S. te Slaa.References
1. Takao H, Hayashi N and Ohtomo K. Effect of scanner in longitudinal studies of brain volume changes. J Magn Reson Imaging. 2011;34:438-44. 2. Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, Pacheco J, Albert M, Killiany R, Blacker D, Maguire P, Rosas D, Makris N, Gollub R, Dale A, Dickerson BC and Fischl B. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009;46:177-92. 3. Heinen R, Bouvy WH, Mendrik AM, Viergever MA, Biessels GJ and de Bresser J. Robustness of Automated Methods for Brain Volume Measurements across Different MRI Field Strengths. PLoS One. 2016;11:e0165719. 4. Steenwijk MD, Amiri H, Schoonheim MM, de Sitter A, Barkhof F, Pouwels PJW and Vrenken H. Agreement of MSmetrix with established methods for measuring cross-sectional and longitudinal brain atrophy. Neuroimage Clin. 2017;15:843-853. 5. Fujimoto K, Robertson TV, Douet V, Garmire D and Stenger VA. A Structurally Anthropomorphic Brain Phantom. Paper presented at: ISMRM; 2015; Toronto, Canada. 6. Chen SJ, Hellier P, Marchal M, Gauvrit JY, Carpentier R, Morandi X and Collins DL. An anthropomorphic polyvinyl alcohol brain phantom based on Colin27 for use in multimodal imaging. Med Phys. 2012;39:554-61. 7. Altermatt A, Santini F, Deligianni X, Magon S, Sprenger T, Kappos L, Cattin P, Wuerfel L and Gaetano L. On the construction of a 3D-printed brain phantom as gold standard for the validation of brain segmentations. Paper presented at: ECRIMS; 2017; Paris, France.Figures
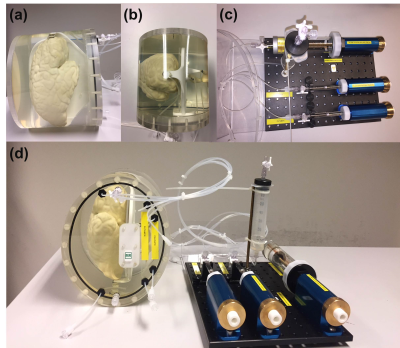
FIG 1. (a) Side-view and (b) top-view of the phantom,
with (c) the volume change system and (d) the complete device. Each structure
had inlet and outlet valves to fill them up with water and to make them
air-free, respectively. Rigid tubes (VYGON) of inner and outer diameter of 1mm
and 2mm, respectively, were used to connect structures to the volume change
system. All structures are mounted in an oval-shaped 3D-printed housing
(PA2200, EOS SLS).
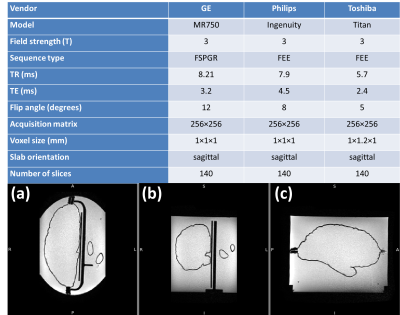
FIG 2. Top panel shows scanners and 3D-T1 sequence
parameters at each scanner. Bottom panel presents MR images of the phantom including
hemisphere, putamen, and caudate nucleus in (a) axial, (b) coronal and (c)
sagittal plane as obtained by Toshiba.
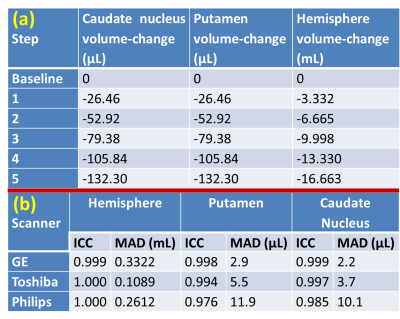
FIG 3. (a) Applied volume
changes of different structures at different steps. (b) Interclass correlation coefficient (ICC)
and mean absolute difference (MAD) for all three structures. Voxel size was 1 µL
for GE and Philips and 1.2 µL for Toshiba.
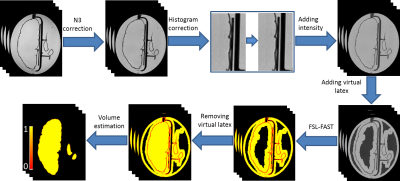
FIG 4.
Diagram of the image analysis pipeline to estimate the volumes of the
hemisphere, putamen and caudate nucleus. Briefly, the software includes
correction for intensity non-uniformity using N3, Gibbs ringing artifact
removal, introduction of an artificial (“virtual”) latex component to balance
the classes in the subsequent FSL-FAST segmentation, and quantification of
total internal volume of each structure from the PVE values obtained from
FSL-FAST.
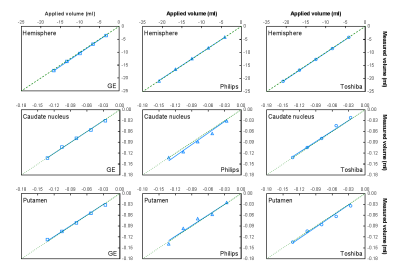
FIG 5. Applied versus measured volume
change at three different scanners for (top row) hemisphere, (middle row)
caudate nucleus, and (bottom row) putamen. The green dotted line represents the
identity line.