3143
A Deep Learning Based Solution for Vertebrae Segmentation of Whole Spine MR Images: A Step Closer to Automated Whole Spine Labeling1GE Healthcare, Bangalore, India
Synopsis
Any reporting on an MR spine scans involves labeling of the vertebrae. Hence, providing labeled spine images for reading can save significant time for radiologists. First step of an automated labeling is reliable segmentation of vertebral bodies. Most of the studies provide methods only for the segmentation and labeling of only a part of the spine. Here, we have used a variant of U-Net based Deep Learning architecture for segmenting vertebrae of Whole Spine. The network was trained with 165 datasets of whole spine images and tested with 8 datasets. We achieved average DICE score of 0.921.
Introduction
MRI is a frequently used modality for imaging spine. Reporting pathology in a spine is always done with reference to specific vertebrae and it involves labeling of each vertebra which is a time-consuming task. Hence, providing labeled spine images for reading can save significant time for radiologists. It is important to have a reliable segmentation of vertebral bodies for successful spine labeling. Recently Deep learning has been employed successfully to various segmentation tasks 1,2,3. Most of the studies, so far, provides methods for the segmentation and labeling of only a part of the spine 4,5. Here, we propose a Deep Learning based method for segmenting vertebrae of Whole Spine.Methods
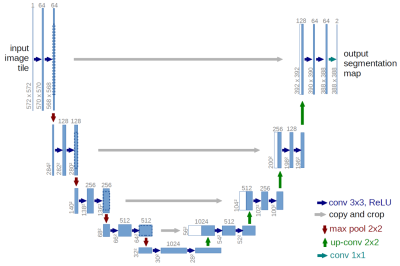
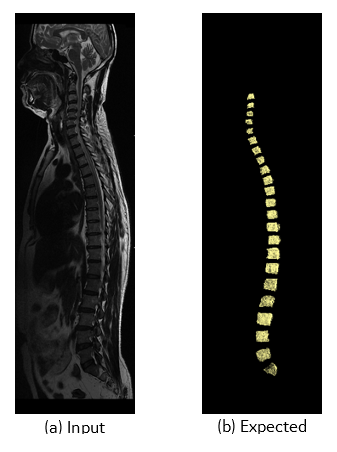
A U-Net (Figure 1) with 4 encoding and 4 decoding layers was selected for this purpose. Each layer consisted of Convolution, ReLU and Max Pooling. In the original UNET implementation 1, pixel wise soft max was used. We modified the loss function in such a way that the product of the ground truth and predicted mask is maximum. The T2 weighted stitched volume from a multi station spine scans performed on GE scanners were used for training along with its 3D segmentation mask (Figure 2). The input and output were resized to 640 x 960 x NoOfSlices. Out of total 183 whole spine datasets 165 were used for training over 10 epochs and 8 were used for testing the trained network. Popular DICE 6 score as follows, DICE= (S∩T)/(S ∪T) where S is the Ground Truth Volume and T is the Deep Network Output Volume, both in terms of number of voxels was used to evaluate the segmentation accuracy.Results
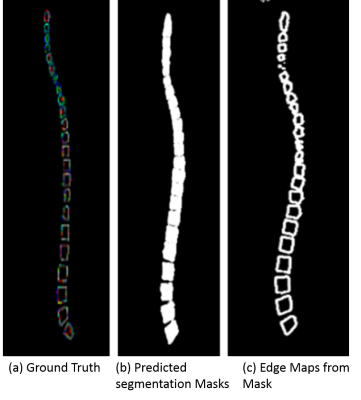
Figure 3 shows an example of one of the test images, comparing the predicted output from the trained modified UNET with the ground truth 3D segmentation mask. The predicted binary masks are also converted into segmentation edge maps based on the prediction values which can be overplayed on the whole spine images for further labeling or planning. The average DICE score on the test data was 0.921. The overall time taken for an experienced user to segment a whole spine was typically around 4 mins ± 30s. In comparison, this proposed automated approach was under 30 seconds in a CPU based inference machine. The overall workflow would all for automated segmentation as well as providing a simple rapid method for the user to review automated findings.Discussion
One of the main advantage of this approach is we are addressing the need of whole spine vertebrae segmentation. Since the ground truth masks are 3D which has information for the entire extent of the vertebrae in the acquired volume, the segmentation accuracy is well improved compared to the deep learning based 2D segmentation techniques. The DICE shows that the method is promising for use for vertebrae segmentation and can be further refined with more datasets to improve accuracy especially in case of datasets with pathologyConclusion
Our work presents a fully automatic segmentation of vertebrae of whole spine MR images trained with 3D segmentation masks, producing significant higher segmentation accuracy. This should eliminate the errors in further vertebrae labeling.Acknowledgements
The authors would like to thank Mr. Ananthakrishna Madhyastha and Sandeep Lakshmipathy, Senior Engineering Managers of GE Healthcare for their insightful comments and suggestionsReferences
1. Ronneberger O., Fischer P., Brox T. (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J., Wells W., Frangi A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham
2. Kalinovsky A. and Kovalev V. Lung image segmentation using Deep Learning methods and convolutional neural networks . In: XIII Int. Conf. on Pattern Recognition and Information Processing, 3-5 October, Minsk, Belarus State University, 2016, pp. 21-24
3. A. Garcia-Garcia, S. Orts-Escolano, S. Oprea, V. Villena-Martinez, and J. Garcia-Rodriguez, “A review on deep learning techniques applied to semantic segmentation,” arXiv preprint arXiv:1704.06857, 2017.
4. C. S. Ling, W. M. Diyana, W. Zaki, A. Hussain and H. A. Hamid, "Semi-automated vertebral segmentation of human spine in MRI images," 2016 International Conference on Advances in Electrical, Electronic and Systems Engineering (ICAEES), Putrajaya, 2016, pp. 120-124
5. Chu C, Belavý DL, Armbrecht G, Bansmann M, Felsenberg D, Zheng G (2015) Fully Automatic Localization and Segmentation of 3D Vertebral Bodies from CT/MR Images via a Learning-Based Method. PLoS ONE10(11): e0143327. https://doi.org/10.1371/journal.pone.0143327
6. Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. doi: 10.2307/1932409.