Na Sang1, Francisco M. Garcia2, Wanshun Wei3, Huabing Li4, Tao Ma1, and Silun Wang1
1YIWEI Medical Inc, Shenzhen, China, 2University of Massachusetts - Amherst, Amherst, MA, United States, 3YIWEI Meidcal Inc, Shenzhen, China, 4ZhongNan University, ChangSha, China
Synopsis
We analyzed the
T1 structural MRI by using deep learning 3D-CNN method. The results indicate that deep learning models
can accurately predict AD patients with diagnostic accuracy of 96%. This can be
achieved using raw MRI data, with a minimum of processing necessary to generate
an accurate AD prediction. Our model shows highly sensitivity and negative
predictive value and thus appropriate for use for screening testing in
population study. Currently model has the potential to be used as a screen biomarker
to investigate the neurodegeneration, brain aging and associated brain
diseases.
Introduction
Alzheimer’s disease (AD) is
the most common form of dementia and its prevalence is set to rise in the
coming decades1. It has been an incredible increase in performance
in classification and regression models mainly sparked by deep learning
techniques. One area in particular that has seen a dramatic improvement in
performance is computer vision, through the use of convolutional neural
networks (CNN) and its variants. We aim to predict the AD with a deep 3D
convolutional neural network (3D-CNN), which can learn generic features
capturing AD biomarkers and adapt to different domain datasets.Methods
Dataset: T1-weighted magnetic
resonance imaging (MRI) data from 90 AD patients with mean age of 71.7 ± 5.9 years and 151 age
and sex matched healthy controls (mean age = 71.4 years ±4.8)
were obtained from the Alzheimer's Disease
Neuroimaging Initiative (ADNI) database. Among them 80% AD patients and controls were recruited
as training dataset and others were regarded as testing dataset. MRI data preprocessing: All MRI data were
preprocessed by using the SPM8 software package (UCL, UK) which included tissue
segmentation; registration and resampling. Each tissue class (i.e., GM and WM)
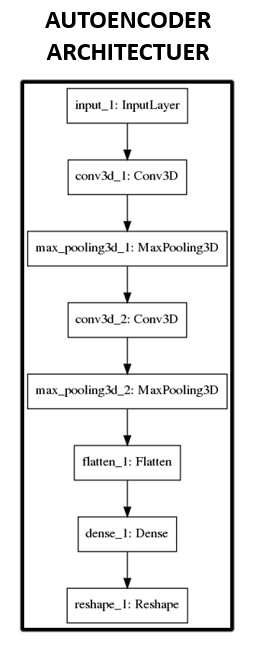
was processed independently after segmentation (Figure1). Feature extraction and classification: Figure 2 and 3 show the
architectures used for the convolutional autoencoder, and the full classifier
neural network. Briefly, given an input image X with height h, width w, and depth d, let us call the dimensionality of X. We
seek an enconder f(X) with
dimensionality k, k << n,
decoder g(f(X)), and train a model
with weights W to minimize the
objective , that is, the square-error between the
decoding of the low dimensional encoding and the original input. Given this new
representation f(X), we train a
neural network classifier using f(X)
as input. Results
Feature
exaction: The autoencoder part of the network is able
to generate low-dimensional representation of the input data, extracting in the
process the features that is most useful to predicted the presence or absence
of the disease. Figure 4 is an example
of the generated 3D images from the autoencoder. Diagnostic accuracy: Our current system shows an average accuracy
of 96% over our testing set doing 5-fold cross validation with sensitivity of
100%, specificity of 92%, positive predictive value of 90% and negative predictive value of 100%. Discussion & Conclusion
Deep learning models with 3D-CNN based on
T1-MRI can accurately predict AD patients. This can be achieved using raw MRI
data, with a minimum of processing necessary to generate an accurate AD prediction. These
estimates of AD prediction model are also significantly heritable, giving external,
genetic, validity to the measure and motivating its use in genetic studies of neurodegeneration
diseases diagnosis. Finally, our model
shows highly sensitivity and negative predictive value and thus appropriate for
use for screening testing in population study. Currently model has the
potential to be used as a screen biomarker to investigate the neurodegeneration,
brain aging and associated brain diseases.Acknowledgements
No acknowledgement found.References
1.
Alzheimer’s Association et al., “2014 alzheimer’s
disease facts and figures,” Alzheimers Dement, vol. 10, no. 2,
pp. e47–e92, 2014.